| Full name: long intergenic non-protein coding RNA 293 | Alias Symbol: BEYLA | ||
| Type: non-coding RNA | Cytoband: 8q11.1 | ||
| Entrez ID: 497634 | HGNC ID: HGNC:39078 | Ensembl Gene: ENSG00000253314 | OMIM ID: 609543 |
| Drug and gene relationship at DGIdb | |||
Expression of LINC00293:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LINC00293 | 497634 | 240682_at | -0.4000 | 0.1004 | |
| GSE26886 | LINC00293 | 497634 | 240682_at | -0.2822 | 0.0452 | |
| GSE45670 | LINC00293 | 497634 | 240682_at | 0.0949 | 0.5459 | |
| GSE53622 | LINC00293 | 497634 | 137908 | 0.6142 | 0.0000 | |
| GSE53624 | LINC00293 | 497634 | 137908 | 0.8874 | 0.0000 | |
| GSE63941 | LINC00293 | 497634 | 240682_at | -0.0139 | 0.9204 | |
| GSE77861 | LINC00293 | 497634 | 240682_at | -0.3650 | 0.0244 |
Upregulated datasets: 0; Downregulated datasets: 0.
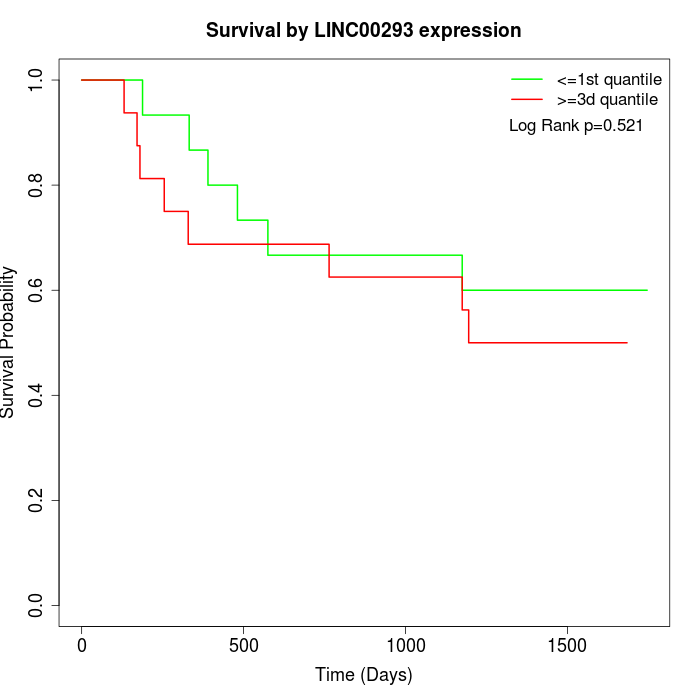
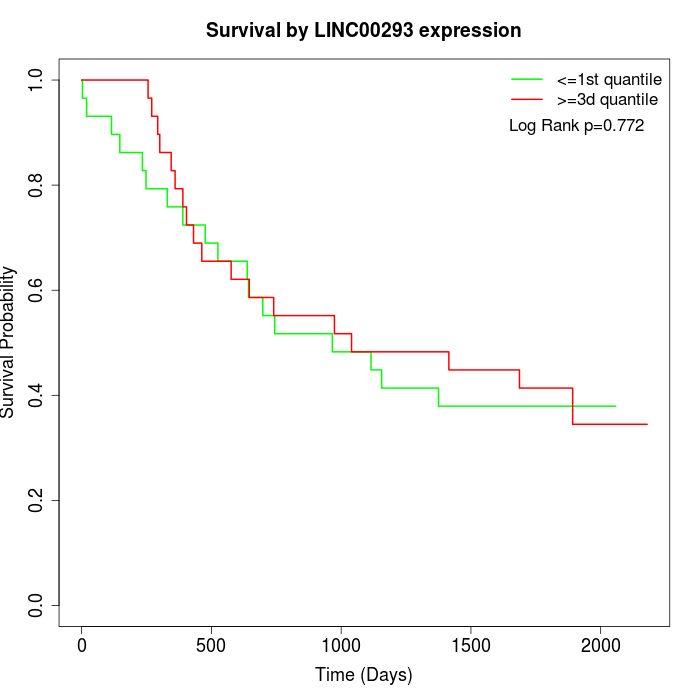
Survival by LINC00293 expression:
Note: Click image to view full size file.
Copy number change of LINC00293:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LINC00293 | 497634 | 16 | 1 | 13 | |
| GSE20123 | LINC00293 | 497634 | 16 | 1 | 13 | |
| GSE43470 | LINC00293 | 497634 | 14 | 3 | 26 | |
| GSE46452 | LINC00293 | 497634 | 22 | 2 | 35 | |
| GSE47630 | LINC00293 | 497634 | 21 | 2 | 17 | |
| GSE54993 | LINC00293 | 497634 | 1 | 16 | 53 | |
| GSE54994 | LINC00293 | 497634 | 27 | 1 | 25 | |
| GSE60625 | LINC00293 | 497634 | 0 | 5 | 6 | |
| GSE74703 | LINC00293 | 497634 | 12 | 2 | 22 | |
| GSE74704 | LINC00293 | 497634 | 12 | 0 | 8 | |
| TCGA | LINC00293 | 497634 | 50 | 4 | 42 |
Total number of gains: 191; Total number of losses: 37; Total Number of normals: 260.
Somatic mutations of LINC00293:
Generating mutation plots.
Highly correlated genes for LINC00293:
Showing top 20/91 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LINC00293 | OR5H1 | 0.710649 | 3 | 0 | 3 |
| LINC00293 | HEMK1 | 0.693634 | 3 | 0 | 3 |
| LINC00293 | FAM95A | 0.692693 | 3 | 0 | 3 |
| LINC00293 | ATP6V1C2 | 0.651549 | 3 | 0 | 3 |
| LINC00293 | PROCA1 | 0.650058 | 3 | 0 | 3 |
| LINC00293 | LINC00687 | 0.647058 | 3 | 0 | 3 |
| LINC00293 | ROM1 | 0.645923 | 3 | 0 | 3 |
| LINC00293 | ZNF280A | 0.636951 | 3 | 0 | 3 |
| LINC00293 | TIAF1 | 0.633387 | 4 | 0 | 4 |
| LINC00293 | CCDC116 | 0.632502 | 3 | 0 | 3 |
| LINC00293 | ABCC8 | 0.62385 | 4 | 0 | 3 |
| LINC00293 | BFSP2-AS1 | 0.618331 | 4 | 0 | 4 |
| LINC00293 | ANKRD18B | 0.616928 | 3 | 0 | 3 |
| LINC00293 | SPATA21 | 0.613553 | 4 | 0 | 4 |
| LINC00293 | SLC25A27 | 0.612943 | 3 | 0 | 3 |
| LINC00293 | SHROOM4 | 0.61069 | 3 | 0 | 3 |
| LINC00293 | LINC00858 | 0.61021 | 3 | 0 | 3 |
| LINC00293 | CD244 | 0.606621 | 4 | 0 | 3 |
| LINC00293 | ZNF667 | 0.605156 | 3 | 0 | 3 |
| LINC00293 | CCDC154 | 0.604841 | 3 | 0 | 3 |
For details and further investigation, click here