| Full name: lipase E, hormone sensitive type | Alias Symbol: HSL | ||
| Type: protein-coding gene | Cytoband: 19q13.2 | ||
| Entrez ID: 3991 | HGNC ID: HGNC:6621 | Ensembl Gene: ENSG00000079435 | OMIM ID: 151750 |
| Drug and gene relationship at DGIdb | |||
LIPE involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04024 | cAMP signaling pathway | |
| hsa04152 | AMPK signaling pathway | |
| hsa04910 | Insulin signaling pathway | |
| hsa04923 | Regulation of lipolysis in adipocytes |
Expression of LIPE:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LIPE | 3991 | 208186_s_at | 0.5366 | 0.3481 | |
| GSE20347 | LIPE | 3991 | 208186_s_at | 0.0261 | 0.8157 | |
| GSE23400 | LIPE | 3991 | 208186_s_at | -0.1166 | 0.0308 | |
| GSE26886 | LIPE | 3991 | 208186_s_at | 0.4614 | 0.0118 | |
| GSE29001 | LIPE | 3991 | 208186_s_at | -0.4837 | 0.0141 | |
| GSE38129 | LIPE | 3991 | 208186_s_at | -0.3232 | 0.1165 | |
| GSE45670 | LIPE | 3991 | 208186_s_at | -0.0491 | 0.7268 | |
| GSE53622 | LIPE | 3991 | 2918 | -0.0395 | 0.5763 | |
| GSE53624 | LIPE | 3991 | 2918 | -0.3729 | 0.0000 | |
| GSE63941 | LIPE | 3991 | 208186_s_at | 0.7620 | 0.0037 | |
| GSE77861 | LIPE | 3991 | 208186_s_at | -0.0539 | 0.8448 | |
| GSE97050 | LIPE | 3991 | A_33_P3294986 | 0.0613 | 0.7727 | |
| SRP007169 | LIPE | 3991 | RNAseq | -2.6443 | 0.0002 | |
| SRP064894 | LIPE | 3991 | RNAseq | -0.7595 | 0.0162 | |
| SRP133303 | LIPE | 3991 | RNAseq | -0.8444 | 0.0016 | |
| SRP159526 | LIPE | 3991 | RNAseq | -0.0042 | 0.9901 | |
| SRP193095 | LIPE | 3991 | RNAseq | -0.2583 | 0.2751 | |
| SRP219564 | LIPE | 3991 | RNAseq | -1.7150 | 0.0000 | |
| TCGA | LIPE | 3991 | RNAseq | -0.0897 | 0.6233 |
Upregulated datasets: 0; Downregulated datasets: 2.
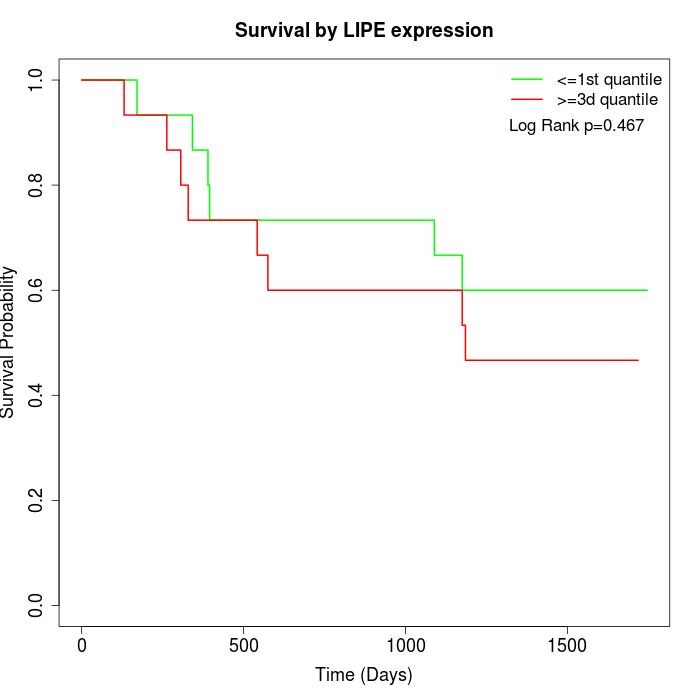
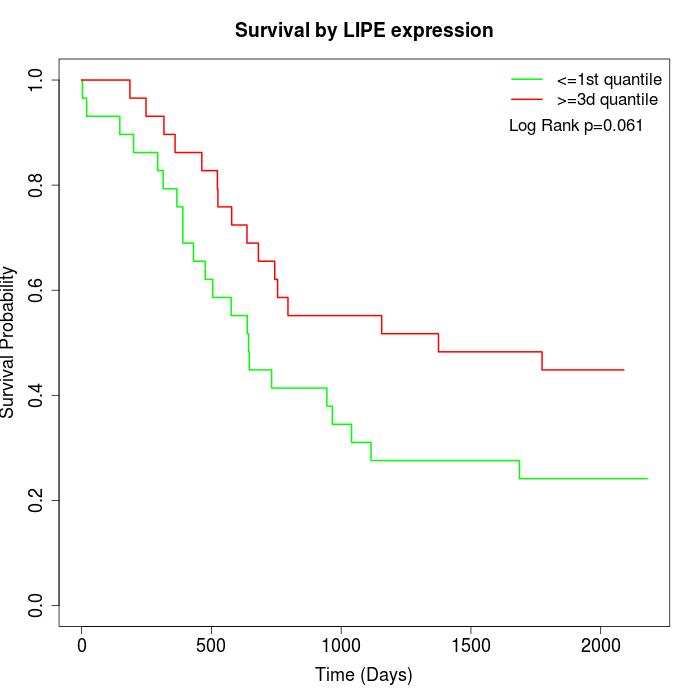
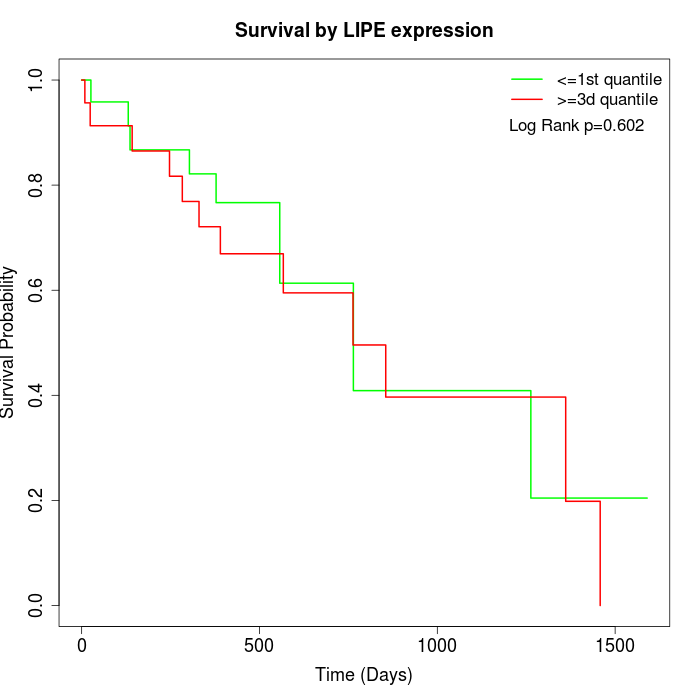
Survival by LIPE expression:
Note: Click image to view full size file.
Copy number change of LIPE:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LIPE | 3991 | 5 | 5 | 20 | |
| GSE20123 | LIPE | 3991 | 4 | 4 | 22 | |
| GSE43470 | LIPE | 3991 | 5 | 11 | 27 | |
| GSE46452 | LIPE | 3991 | 47 | 1 | 11 | |
| GSE47630 | LIPE | 3991 | 8 | 6 | 26 | |
| GSE54993 | LIPE | 3991 | 17 | 4 | 49 | |
| GSE54994 | LIPE | 3991 | 7 | 12 | 34 | |
| GSE60625 | LIPE | 3991 | 9 | 0 | 2 | |
| GSE74703 | LIPE | 3991 | 5 | 7 | 24 | |
| GSE74704 | LIPE | 3991 | 4 | 2 | 14 | |
| TCGA | LIPE | 3991 | 13 | 16 | 67 |
Total number of gains: 124; Total number of losses: 68; Total Number of normals: 296.
Somatic mutations of LIPE:
Generating mutation plots.
Highly correlated genes for LIPE:
Showing top 20/372 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LIPE | FAM189B | 0.802272 | 3 | 0 | 3 |
| LIPE | ZSWIM3 | 0.756402 | 3 | 0 | 3 |
| LIPE | ARHGDIA | 0.752097 | 3 | 0 | 3 |
| LIPE | DYDC2 | 0.738757 | 3 | 0 | 3 |
| LIPE | CSNK1E | 0.734277 | 3 | 0 | 3 |
| LIPE | ZNF687 | 0.732803 | 4 | 0 | 4 |
| LIPE | PYGO2 | 0.726183 | 3 | 0 | 3 |
| LIPE | GNB3 | 0.723532 | 3 | 0 | 3 |
| LIPE | PPP2R5D | 0.721278 | 3 | 0 | 3 |
| LIPE | GRIPAP1 | 0.715938 | 3 | 0 | 3 |
| LIPE | MIIP | 0.714984 | 4 | 0 | 3 |
| LIPE | FAM53A | 0.709822 | 3 | 0 | 3 |
| LIPE | PRR15 | 0.707965 | 3 | 0 | 3 |
| LIPE | TMEM151A | 0.700418 | 4 | 0 | 3 |
| LIPE | TAOK2 | 0.69995 | 5 | 0 | 5 |
| LIPE | CABP4 | 0.69477 | 4 | 0 | 3 |
| LIPE | CNTD2 | 0.693528 | 4 | 0 | 4 |
| LIPE | OPTC | 0.691409 | 3 | 0 | 3 |
| LIPE | SNORA71A | 0.691006 | 3 | 0 | 3 |
| LIPE | ACSM5 | 0.688945 | 4 | 0 | 4 |
For details and further investigation, click here