| Full name: phosphatidylinositol glycan anchor biosynthesis class Q | Alias Symbol: hGPI1|GPI1 | ||
| Type: protein-coding gene | Cytoband: 16p13.3 | ||
| Entrez ID: 9091 | HGNC ID: HGNC:14135 | Ensembl Gene: ENSG00000007541 | OMIM ID: 605754 |
| Drug and gene relationship at DGIdb | |||
Expression of PIGQ:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PIGQ | 9091 | 1555359_at | 0.1326 | 0.6318 | |
| GSE20347 | PIGQ | 9091 | 204144_s_at | 0.0777 | 0.3207 | |
| GSE23400 | PIGQ | 9091 | 204144_s_at | -0.0299 | 0.4532 | |
| GSE26886 | PIGQ | 9091 | 1555359_at | -0.0130 | 0.9494 | |
| GSE29001 | PIGQ | 9091 | 204144_s_at | -0.0550 | 0.6719 | |
| GSE38129 | PIGQ | 9091 | 204144_s_at | 0.1119 | 0.1454 | |
| GSE45670 | PIGQ | 9091 | 1555359_at | 0.0476 | 0.6559 | |
| GSE53622 | PIGQ | 9091 | 98776 | 0.0247 | 0.7709 | |
| GSE53624 | PIGQ | 9091 | 49409 | 0.4459 | 0.0000 | |
| GSE63941 | PIGQ | 9091 | 1555359_at | 0.0603 | 0.7222 | |
| GSE77861 | PIGQ | 9091 | 1555359_at | 0.0099 | 0.9447 | |
| GSE97050 | PIGQ | 9091 | A_33_P3215178 | -0.0897 | 0.6893 | |
| SRP007169 | PIGQ | 9091 | RNAseq | -0.5528 | 0.1669 | |
| SRP008496 | PIGQ | 9091 | RNAseq | -0.5260 | 0.0748 | |
| SRP064894 | PIGQ | 9091 | RNAseq | 0.5446 | 0.0856 | |
| SRP133303 | PIGQ | 9091 | RNAseq | 0.2496 | 0.1973 | |
| SRP159526 | PIGQ | 9091 | RNAseq | 0.4342 | 0.0845 | |
| SRP193095 | PIGQ | 9091 | RNAseq | 0.4013 | 0.0004 | |
| SRP219564 | PIGQ | 9091 | RNAseq | 0.2043 | 0.5360 | |
| TCGA | PIGQ | 9091 | RNAseq | -0.0212 | 0.6715 |
Upregulated datasets: 0; Downregulated datasets: 0.
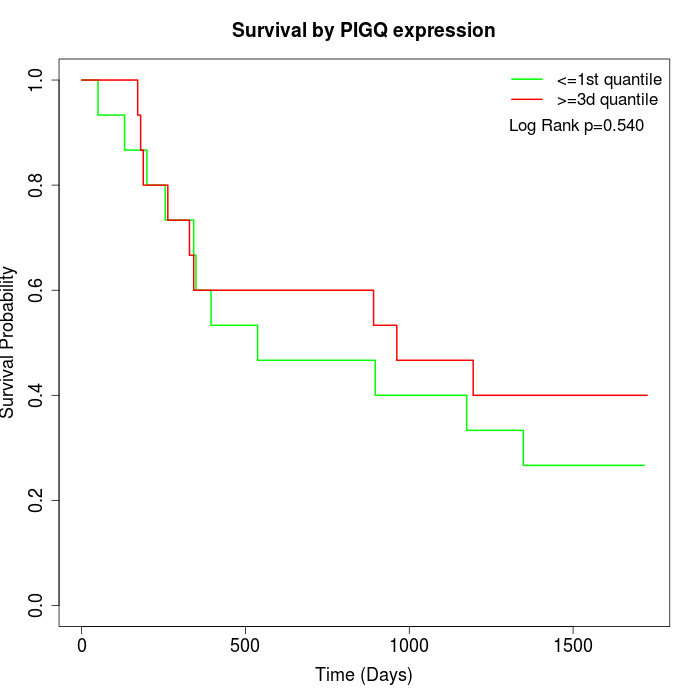
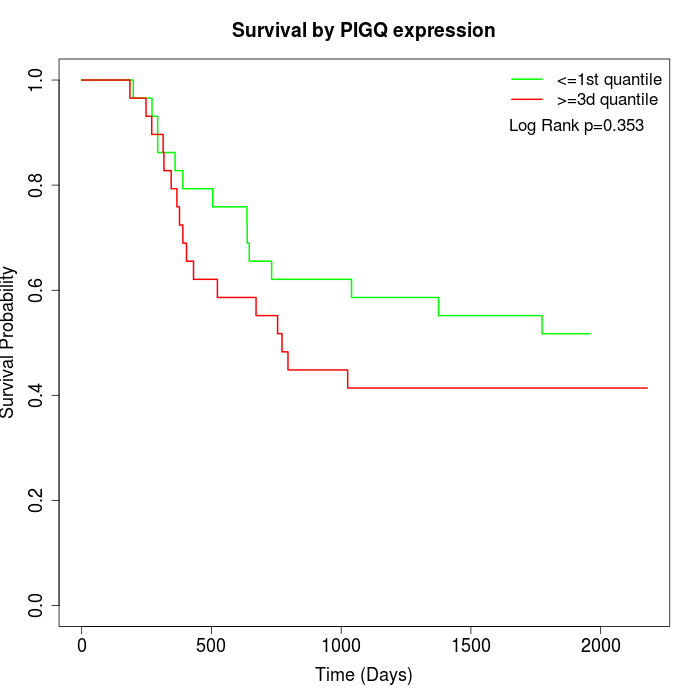
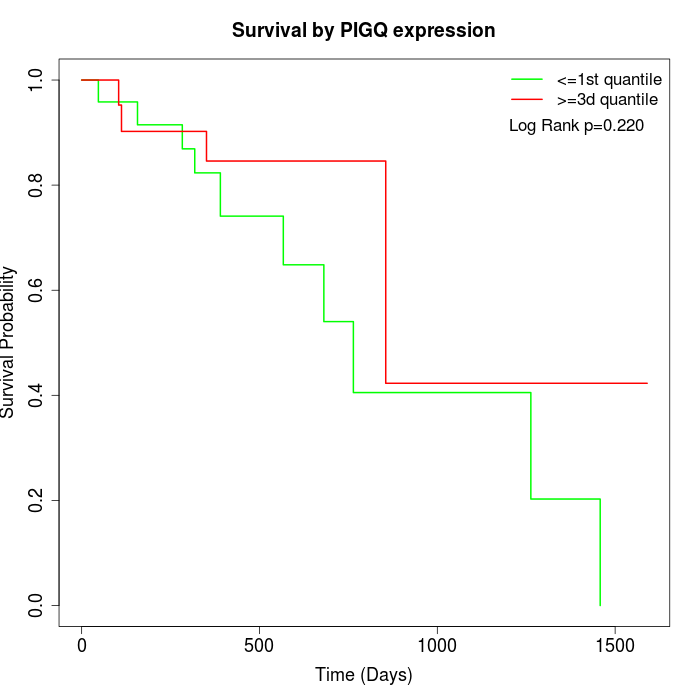
Survival by PIGQ expression:
Note: Click image to view full size file.
Copy number change of PIGQ:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PIGQ | 9091 | 5 | 5 | 20 | |
| GSE20123 | PIGQ | 9091 | 5 | 4 | 21 | |
| GSE43470 | PIGQ | 9091 | 4 | 7 | 32 | |
| GSE46452 | PIGQ | 9091 | 38 | 1 | 20 | |
| GSE47630 | PIGQ | 9091 | 13 | 6 | 21 | |
| GSE54993 | PIGQ | 9091 | 3 | 5 | 62 | |
| GSE54994 | PIGQ | 9091 | 4 | 10 | 39 | |
| GSE60625 | PIGQ | 9091 | 4 | 0 | 7 | |
| GSE74703 | PIGQ | 9091 | 4 | 5 | 27 | |
| GSE74704 | PIGQ | 9091 | 3 | 2 | 15 | |
| TCGA | PIGQ | 9091 | 20 | 15 | 61 |
Total number of gains: 103; Total number of losses: 60; Total Number of normals: 325.
Somatic mutations of PIGQ:
Generating mutation plots.
Highly correlated genes for PIGQ:
Showing top 20/210 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PIGQ | ZNF687 | 0.737845 | 4 | 0 | 4 |
| PIGQ | C7orf26 | 0.722512 | 3 | 0 | 3 |
| PIGQ | FAM181A | 0.720559 | 3 | 0 | 3 |
| PIGQ | LYPD6 | 0.719885 | 3 | 0 | 3 |
| PIGQ | GRM4 | 0.719769 | 3 | 0 | 3 |
| PIGQ | POM121L12 | 0.714778 | 3 | 0 | 3 |
| PIGQ | C2orf81 | 0.710455 | 3 | 0 | 3 |
| PIGQ | BPIFB4 | 0.707201 | 4 | 0 | 4 |
| PIGQ | ZNF639 | 0.704646 | 3 | 0 | 3 |
| PIGQ | MOGAT3 | 0.702529 | 3 | 0 | 3 |
| PIGQ | GMNC | 0.686505 | 3 | 0 | 3 |
| PIGQ | TEX13A | 0.681154 | 3 | 0 | 3 |
| PIGQ | SYPL2 | 0.678603 | 4 | 0 | 3 |
| PIGQ | KCNIP2-AS1 | 0.671981 | 3 | 0 | 3 |
| PIGQ | ZNF236 | 0.67129 | 4 | 0 | 3 |
| PIGQ | TSPAN32 | 0.670527 | 3 | 0 | 3 |
| PIGQ | HDGFL1 | 0.669348 | 3 | 0 | 3 |
| PIGQ | PPP5D1 | 0.664595 | 3 | 0 | 3 |
| PIGQ | GNAL | 0.663636 | 3 | 0 | 3 |
| PIGQ | INSM2 | 0.661397 | 4 | 0 | 3 |
For details and further investigation, click here