| Full name: LY6/PLAUR domain containing 6 | Alias Symbol: MGC52057 | ||
| Type: protein-coding gene | Cytoband: 2q23.2 | ||
| Entrez ID: 130574 | HGNC ID: HGNC:28751 | Ensembl Gene: ENSG00000187123 | OMIM ID: 613359 |
| Drug and gene relationship at DGIdb | |||
Expression of LYPD6:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LYPD6 | 130574 | 227764_at | 0.3841 | 0.6393 | |
| GSE26886 | LYPD6 | 130574 | 227764_at | 0.6565 | 0.1594 | |
| GSE45670 | LYPD6 | 130574 | 227764_at | 0.1443 | 0.5609 | |
| GSE53622 | LYPD6 | 130574 | 107465 | 0.4530 | 0.0001 | |
| GSE53624 | LYPD6 | 130574 | 107465 | 0.4814 | 0.0000 | |
| GSE63941 | LYPD6 | 130574 | 227764_at | 0.7432 | 0.3136 | |
| GSE77861 | LYPD6 | 130574 | 227764_at | 0.0899 | 0.7152 | |
| GSE97050 | LYPD6 | 130574 | A_33_P3359047 | 0.0007 | 0.9975 | |
| SRP133303 | LYPD6 | 130574 | RNAseq | 1.1085 | 0.0409 | |
| SRP219564 | LYPD6 | 130574 | RNAseq | 0.7231 | 0.1989 | |
| TCGA | LYPD6 | 130574 | RNAseq | -0.1890 | 0.5113 |
Upregulated datasets: 1; Downregulated datasets: 0.
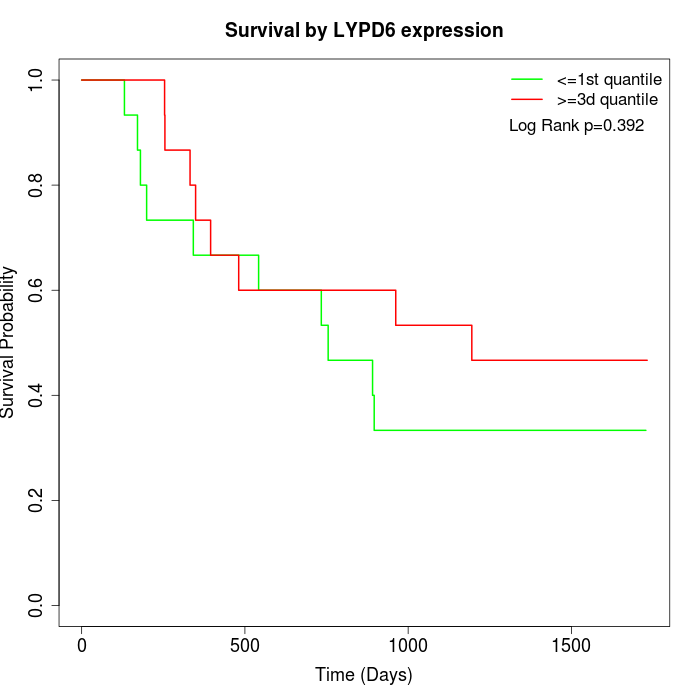
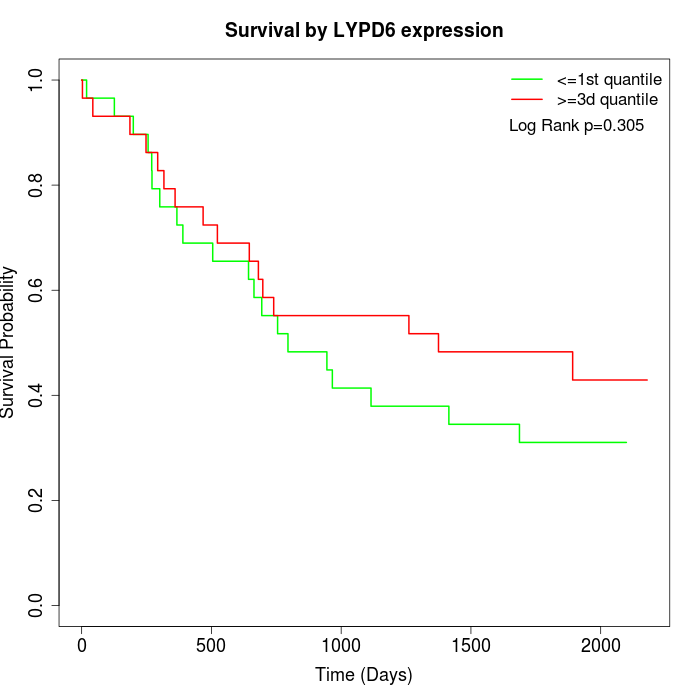
Survival by LYPD6 expression:
Note: Click image to view full size file.
Copy number change of LYPD6:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LYPD6 | 130574 | 3 | 3 | 24 | |
| GSE20123 | LYPD6 | 130574 | 3 | 3 | 24 | |
| GSE43470 | LYPD6 | 130574 | 3 | 1 | 39 | |
| GSE46452 | LYPD6 | 130574 | 1 | 4 | 54 | |
| GSE47630 | LYPD6 | 130574 | 5 | 3 | 32 | |
| GSE54993 | LYPD6 | 130574 | 0 | 5 | 65 | |
| GSE54994 | LYPD6 | 130574 | 11 | 2 | 40 | |
| GSE60625 | LYPD6 | 130574 | 0 | 3 | 8 | |
| GSE74703 | LYPD6 | 130574 | 2 | 1 | 33 | |
| GSE74704 | LYPD6 | 130574 | 3 | 1 | 16 | |
| TCGA | LYPD6 | 130574 | 21 | 8 | 67 |
Total number of gains: 52; Total number of losses: 34; Total Number of normals: 402.
Somatic mutations of LYPD6:
Generating mutation plots.
Highly correlated genes for LYPD6:
Showing top 20/34 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LYPD6 | SCN9A | 0.738182 | 3 | 0 | 3 |
| LYPD6 | VWDE | 0.723598 | 3 | 0 | 3 |
| LYPD6 | PIGQ | 0.719885 | 3 | 0 | 3 |
| LYPD6 | LAYN | 0.700106 | 3 | 0 | 3 |
| LYPD6 | DUSP9 | 0.680173 | 3 | 0 | 3 |
| LYPD6 | MPP3 | 0.677269 | 4 | 0 | 3 |
| LYPD6 | CUL4A | 0.666605 | 3 | 0 | 3 |
| LYPD6 | DSCAM | 0.657883 | 3 | 0 | 3 |
| LYPD6 | CDR2L | 0.651182 | 4 | 0 | 3 |
| LYPD6 | RANGRF | 0.650501 | 3 | 0 | 3 |
| LYPD6 | DCUN1D1 | 0.64703 | 4 | 0 | 3 |
| LYPD6 | BHMT | 0.643247 | 3 | 0 | 3 |
| LYPD6 | KATNAL2 | 0.636893 | 3 | 0 | 3 |
| LYPD6 | DLX6-AS1 | 0.630395 | 3 | 0 | 3 |
| LYPD6 | HOXD-AS2 | 0.624993 | 3 | 0 | 3 |
| LYPD6 | RANBP17 | 0.619139 | 4 | 0 | 4 |
| LYPD6 | ZFHX4 | 0.614181 | 3 | 0 | 3 |
| LYPD6 | ZNF639 | 0.612493 | 5 | 0 | 3 |
| LYPD6 | SMKR1 | 0.60971 | 3 | 0 | 3 |
| LYPD6 | CHST10 | 0.608869 | 5 | 0 | 3 |
For details and further investigation, click here