| Full name: INSM transcriptional repressor 2 | Alias Symbol: IA-6|Mlt1 | ||
| Type: protein-coding gene | Cytoband: 14q13.2 | ||
| Entrez ID: 84684 | HGNC ID: HGNC:17539 | Ensembl Gene: ENSG00000168348 | OMIM ID: 614027 |
| Drug and gene relationship at DGIdb | |||
Expression of INSM2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | INSM2 | 84684 | 240841_at | 0.1347 | 0.7364 | |
| GSE26886 | INSM2 | 84684 | 240841_at | 0.1677 | 0.1751 | |
| GSE45670 | INSM2 | 84684 | 240841_at | 0.1049 | 0.1421 | |
| GSE53622 | INSM2 | 84684 | 39111 | 0.2143 | 0.1981 | |
| GSE53624 | INSM2 | 84684 | 39111 | -0.0721 | 0.4868 | |
| GSE63941 | INSM2 | 84684 | 240841_at | 0.3361 | 0.0229 | |
| GSE77861 | INSM2 | 84684 | 240841_at | 0.0425 | 0.6901 | |
| TCGA | INSM2 | 84684 | RNAseq | 0.2406 | 0.7925 |
Upregulated datasets: 0; Downregulated datasets: 0.
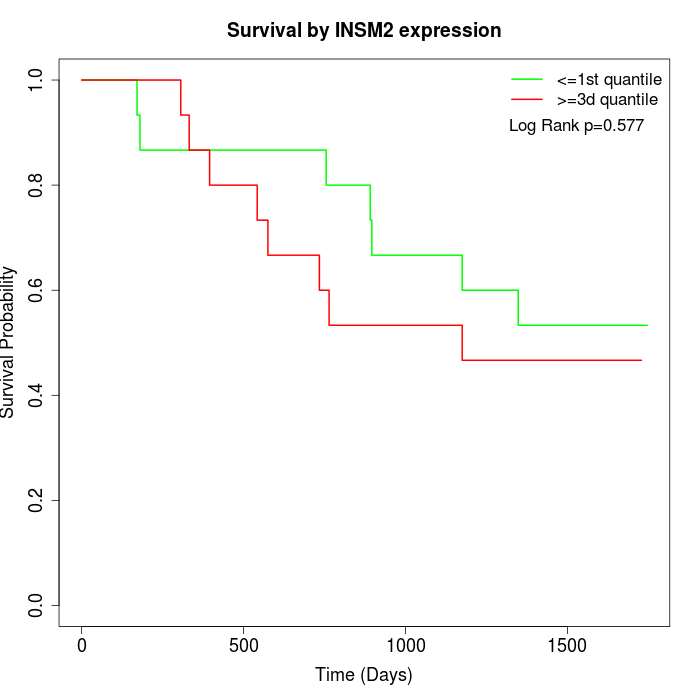
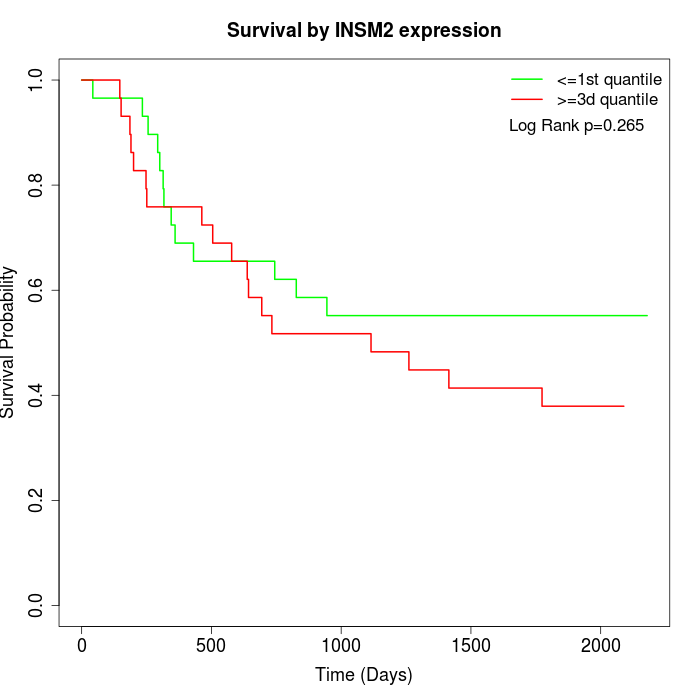
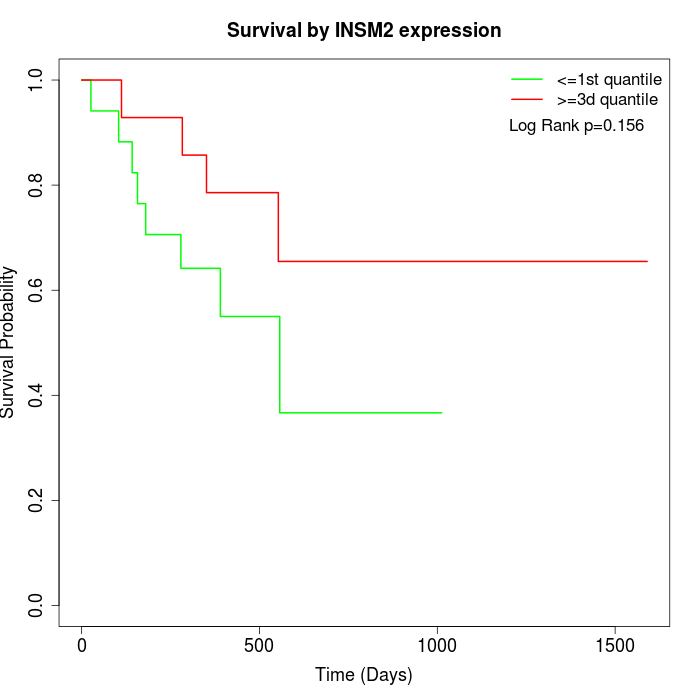
Survival by INSM2 expression:
Note: Click image to view full size file.
Copy number change of INSM2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | INSM2 | 84684 | 10 | 4 | 16 | |
| GSE20123 | INSM2 | 84684 | 11 | 4 | 15 | |
| GSE43470 | INSM2 | 84684 | 9 | 3 | 31 | |
| GSE46452 | INSM2 | 84684 | 18 | 2 | 39 | |
| GSE47630 | INSM2 | 84684 | 11 | 10 | 19 | |
| GSE54993 | INSM2 | 84684 | 3 | 10 | 57 | |
| GSE54994 | INSM2 | 84684 | 19 | 5 | 29 | |
| GSE60625 | INSM2 | 84684 | 0 | 2 | 9 | |
| GSE74703 | INSM2 | 84684 | 8 | 3 | 25 | |
| GSE74704 | INSM2 | 84684 | 5 | 3 | 12 | |
| TCGA | INSM2 | 84684 | 34 | 14 | 48 |
Total number of gains: 128; Total number of losses: 60; Total Number of normals: 300.
Somatic mutations of INSM2:
Generating mutation plots.
Highly correlated genes for INSM2:
Showing top 20/83 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| INSM2 | ENAM | 0.726118 | 3 | 0 | 3 |
| INSM2 | KRBA1 | 0.704568 | 3 | 0 | 3 |
| INSM2 | PIGQ | 0.661397 | 4 | 0 | 3 |
| INSM2 | BCL9L | 0.656556 | 3 | 0 | 3 |
| INSM2 | TLL2 | 0.649413 | 3 | 0 | 3 |
| INSM2 | PRAMEF11 | 0.647073 | 4 | 0 | 3 |
| INSM2 | IRF2BP1 | 0.637559 | 4 | 0 | 4 |
| INSM2 | SIK1 | 0.637076 | 3 | 0 | 3 |
| INSM2 | NR4A1 | 0.632363 | 3 | 0 | 3 |
| INSM2 | RILP | 0.630942 | 3 | 0 | 3 |
| INSM2 | LRRC10B | 0.625022 | 3 | 0 | 3 |
| INSM2 | ERF | 0.623425 | 3 | 0 | 3 |
| INSM2 | DNAH11 | 0.623275 | 3 | 0 | 3 |
| INSM2 | AAAS | 0.622188 | 3 | 0 | 3 |
| INSM2 | PNPLA2 | 0.619166 | 4 | 0 | 3 |
| INSM2 | SNAPC2 | 0.617336 | 3 | 0 | 3 |
| INSM2 | MPV17L2 | 0.61577 | 3 | 0 | 3 |
| INSM2 | C14orf178 | 0.615305 | 4 | 0 | 3 |
| INSM2 | GBX1 | 0.615243 | 3 | 0 | 3 |
| INSM2 | RAB3IL1 | 0.610716 | 3 | 0 | 3 |
For details and further investigation, click here