| Full name: protein kinase AMP-activated non-catalytic subunit gamma 3 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 2q35 | ||
| Entrez ID: 53632 | HGNC ID: HGNC:9387 | Ensembl Gene: ENSG00000115592 | OMIM ID: 604976 |
| Drug and gene relationship at DGIdb | |||
PRKAG3 involved pathways:
Expression of PRKAG3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PRKAG3 | 53632 | 223904_at | -0.1125 | 0.5444 | |
| GSE26886 | PRKAG3 | 53632 | 223904_at | 0.0798 | 0.5081 | |
| GSE45670 | PRKAG3 | 53632 | 223904_at | 0.0498 | 0.5747 | |
| GSE53622 | PRKAG3 | 53632 | 105471 | 0.5050 | 0.0000 | |
| GSE53624 | PRKAG3 | 53632 | 105471 | 0.4378 | 0.0000 | |
| GSE63941 | PRKAG3 | 53632 | 223904_at | 0.0660 | 0.7105 | |
| GSE77861 | PRKAG3 | 53632 | 223904_at | -0.0143 | 0.9219 | |
| GSE97050 | PRKAG3 | 53632 | A_33_P3244843 | -0.0801 | 0.8207 | |
| TCGA | PRKAG3 | 53632 | RNAseq | 1.3592 | 0.1824 |
Upregulated datasets: 0; Downregulated datasets: 0.
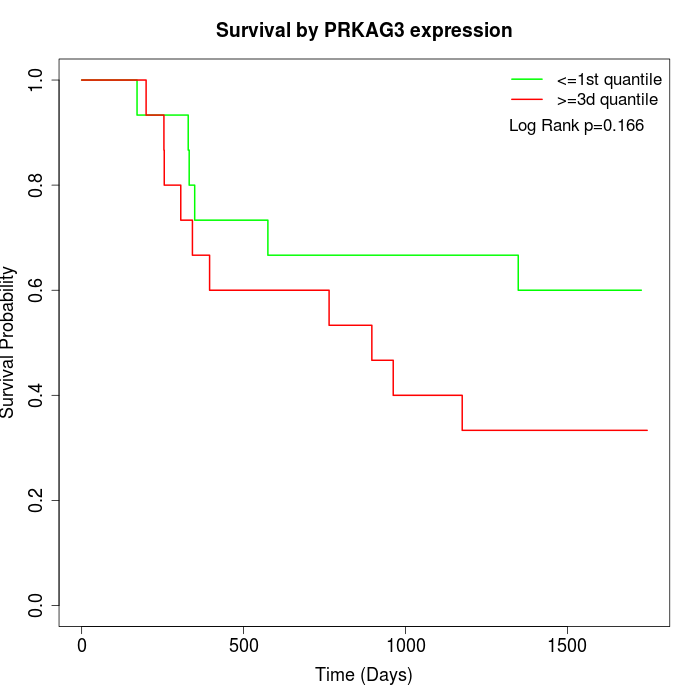
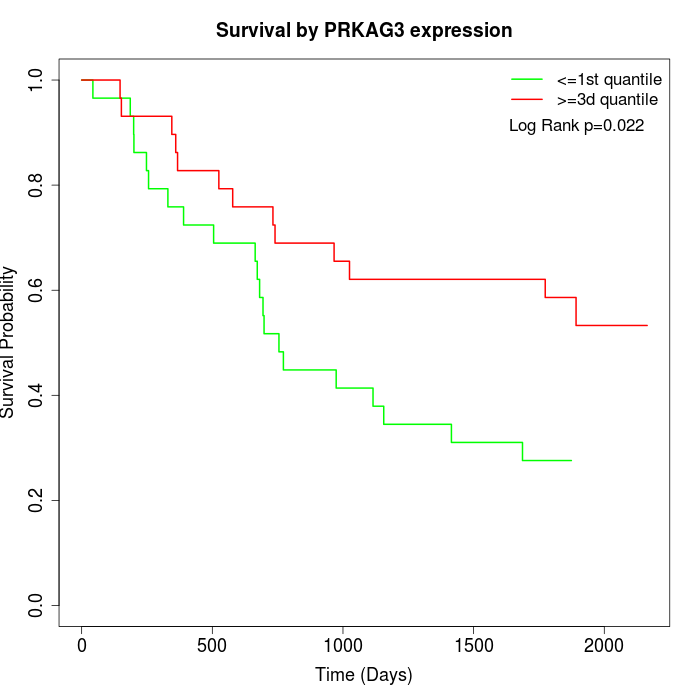
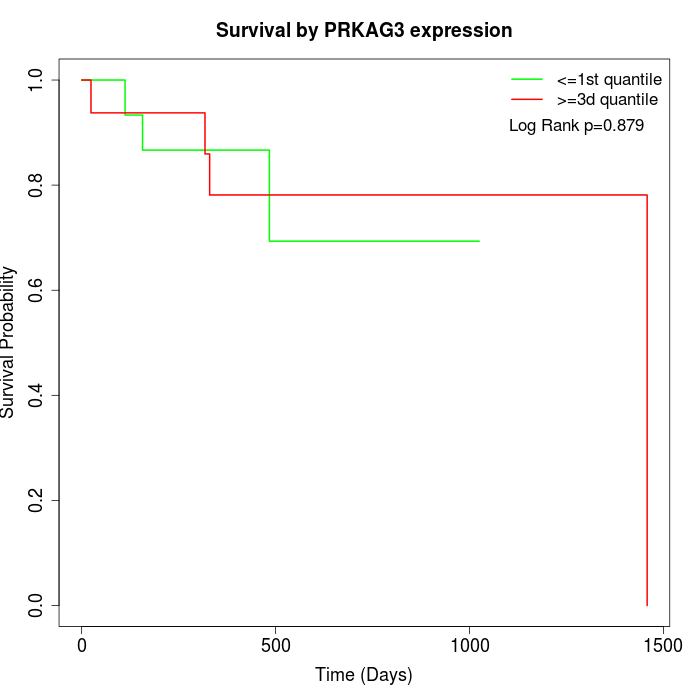
Survival by PRKAG3 expression:
Note: Click image to view full size file.
Copy number change of PRKAG3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PRKAG3 | 53632 | 1 | 14 | 15 | |
| GSE20123 | PRKAG3 | 53632 | 1 | 13 | 16 | |
| GSE43470 | PRKAG3 | 53632 | 1 | 7 | 35 | |
| GSE46452 | PRKAG3 | 53632 | 0 | 5 | 54 | |
| GSE47630 | PRKAG3 | 53632 | 4 | 5 | 31 | |
| GSE54993 | PRKAG3 | 53632 | 1 | 2 | 67 | |
| GSE54994 | PRKAG3 | 53632 | 7 | 10 | 36 | |
| GSE60625 | PRKAG3 | 53632 | 0 | 3 | 8 | |
| GSE74703 | PRKAG3 | 53632 | 1 | 5 | 30 | |
| GSE74704 | PRKAG3 | 53632 | 1 | 7 | 12 | |
| TCGA | PRKAG3 | 53632 | 11 | 27 | 58 |
Total number of gains: 28; Total number of losses: 98; Total Number of normals: 362.
Somatic mutations of PRKAG3:
Generating mutation plots.
Highly correlated genes for PRKAG3:
Showing top 20/151 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PRKAG3 | MRGPRG | 0.776075 | 3 | 0 | 3 |
| PRKAG3 | TMEM235 | 0.75171 | 3 | 0 | 3 |
| PRKAG3 | SLC6A3 | 0.731834 | 3 | 0 | 3 |
| PRKAG3 | PRLH | 0.729098 | 3 | 0 | 3 |
| PRKAG3 | GCLM | 0.724461 | 3 | 0 | 3 |
| PRKAG3 | C2CD4D | 0.718677 | 3 | 0 | 3 |
| PRKAG3 | BLACE | 0.715892 | 3 | 0 | 3 |
| PRKAG3 | CHD5 | 0.711827 | 4 | 0 | 4 |
| PRKAG3 | DCDC1 | 0.709551 | 3 | 0 | 3 |
| PRKAG3 | C9orf153 | 0.707725 | 3 | 0 | 3 |
| PRKAG3 | GRIK4 | 0.705818 | 3 | 0 | 3 |
| PRKAG3 | MS4A18 | 0.704844 | 3 | 0 | 3 |
| PRKAG3 | POM121L12 | 0.697672 | 3 | 0 | 3 |
| PRKAG3 | USH1C | 0.697663 | 3 | 0 | 3 |
| PRKAG3 | CIB3 | 0.697146 | 4 | 0 | 4 |
| PRKAG3 | DNAI1 | 0.696966 | 3 | 0 | 3 |
| PRKAG3 | SCN9A | 0.696806 | 3 | 0 | 3 |
| PRKAG3 | C19orf71 | 0.695462 | 3 | 0 | 3 |
| PRKAG3 | RIMS4 | 0.692029 | 3 | 0 | 3 |
| PRKAG3 | ONECUT1 | 0.689343 | 3 | 0 | 3 |
For details and further investigation, click here