| Full name: ring finger protein 133 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 7q31.32 | ||
| Entrez ID: 168433 | HGNC ID: HGNC:21154 | Ensembl Gene: ENSG00000188050 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of RNF133:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RNF133 | 168433 | 231373_at | -0.0268 | 0.9361 | |
| GSE26886 | RNF133 | 168433 | 231373_at | -0.0006 | 0.9948 | |
| GSE45670 | RNF133 | 168433 | 231373_at | 0.0494 | 0.6015 | |
| GSE53622 | RNF133 | 168433 | 33636 | -0.0559 | 0.7226 | |
| GSE53624 | RNF133 | 168433 | 33636 | -0.0874 | 0.3915 | |
| GSE63941 | RNF133 | 168433 | 231373_at | 0.1619 | 0.2151 | |
| GSE77861 | RNF133 | 168433 | 231373_at | 0.0393 | 0.6739 | |
| TCGA | RNF133 | 168433 | RNAseq | -1.6093 | 0.0285 |
Upregulated datasets: 0; Downregulated datasets: 1.
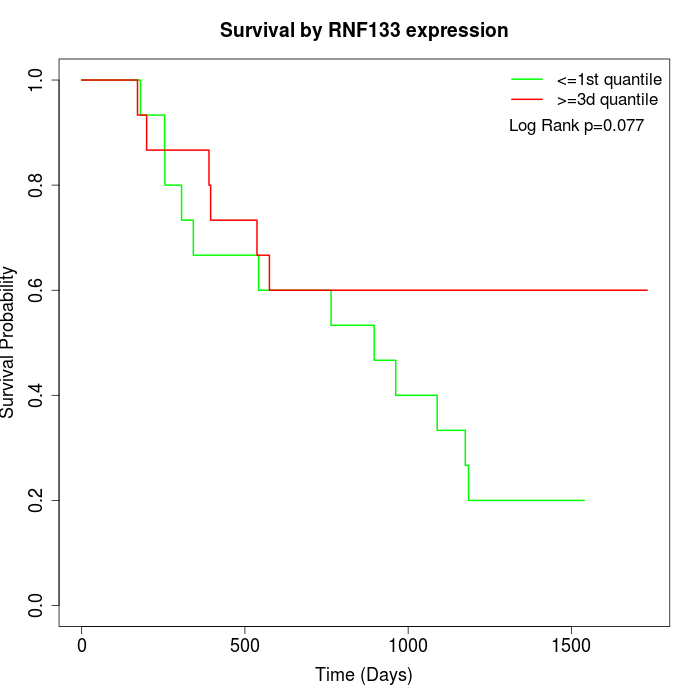
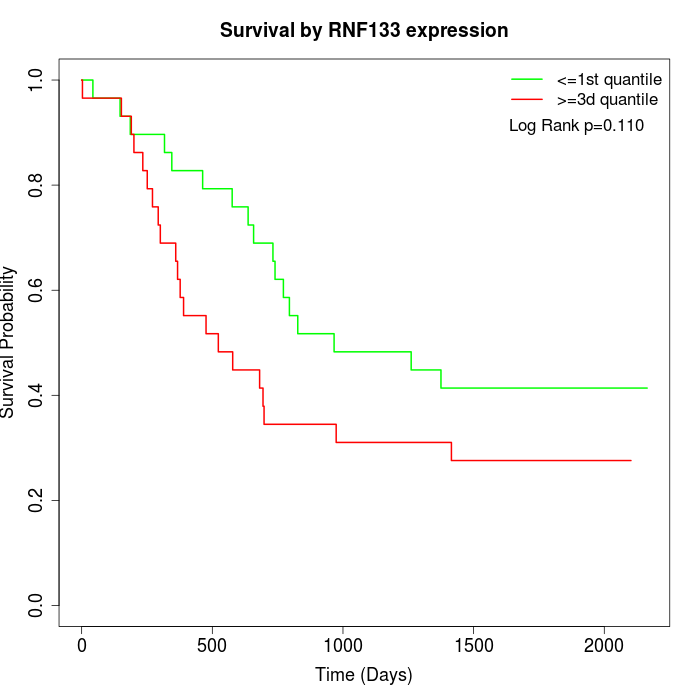
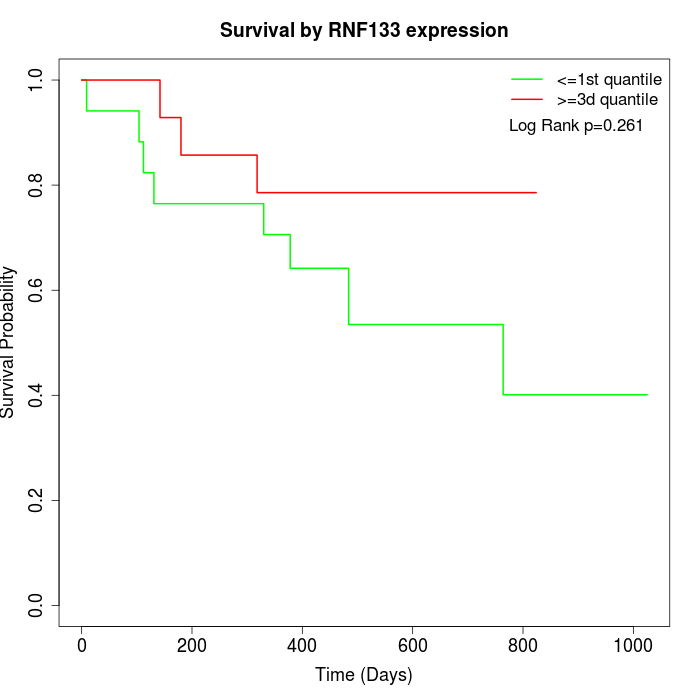
Survival by RNF133 expression:
Note: Click image to view full size file.
Copy number change of RNF133:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RNF133 | 168433 | 7 | 2 | 21 | |
| GSE20123 | RNF133 | 168433 | 8 | 2 | 20 | |
| GSE43470 | RNF133 | 168433 | 2 | 4 | 37 | |
| GSE46452 | RNF133 | 168433 | 8 | 1 | 50 | |
| GSE47630 | RNF133 | 168433 | 7 | 4 | 29 | |
| GSE54993 | RNF133 | 168433 | 3 | 5 | 62 | |
| GSE54994 | RNF133 | 168433 | 10 | 6 | 37 | |
| GSE60625 | RNF133 | 168433 | 0 | 0 | 11 | |
| GSE74703 | RNF133 | 168433 | 2 | 4 | 30 | |
| GSE74704 | RNF133 | 168433 | 5 | 2 | 13 | |
| TCGA | RNF133 | 168433 | 27 | 21 | 48 |
Total number of gains: 79; Total number of losses: 51; Total Number of normals: 358.
Somatic mutations of RNF133:
Generating mutation plots.
Highly correlated genes for RNF133:
Showing top 20/31 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RNF133 | ACSM1 | 0.751046 | 3 | 0 | 3 |
| RNF133 | ADAP1 | 0.682811 | 3 | 0 | 3 |
| RNF133 | SNX32 | 0.633954 | 4 | 0 | 3 |
| RNF133 | HNF1B | 0.62518 | 3 | 0 | 3 |
| RNF133 | CLDN14 | 0.609668 | 4 | 0 | 3 |
| RNF133 | CLYBL-AS2 | 0.606844 | 3 | 0 | 3 |
| RNF133 | ZSCAN10 | 0.600019 | 3 | 0 | 3 |
| RNF133 | KLHL11 | 0.599192 | 3 | 0 | 3 |
| RNF133 | ITIH6 | 0.586802 | 3 | 0 | 3 |
| RNF133 | B4GALNT1 | 0.585283 | 3 | 0 | 3 |
| RNF133 | MAPT-AS1 | 0.582553 | 4 | 0 | 3 |
| RNF133 | MYOD1 | 0.576454 | 4 | 0 | 3 |
| RNF133 | GRIK5 | 0.57636 | 5 | 0 | 3 |
| RNF133 | TSACC | 0.566843 | 3 | 0 | 3 |
| RNF133 | SLC5A2 | 0.566515 | 4 | 0 | 3 |
| RNF133 | NNAT | 0.559908 | 3 | 0 | 3 |
| RNF133 | CEACAM4 | 0.550529 | 3 | 0 | 3 |
| RNF133 | SMCP | 0.53856 | 4 | 0 | 3 |
| RNF133 | TEX13B | 0.538135 | 5 | 0 | 3 |
| RNF133 | LONRF3 | 0.538032 | 3 | 0 | 3 |
For details and further investigation, click here