| Full name: zinc finger and SCAN domain containing 10 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 16p13.3 | ||
| Entrez ID: 84891 | HGNC ID: HGNC:12997 | Ensembl Gene: ENSG00000130182 | OMIM ID: 618365 |
| Drug and gene relationship at DGIdb | |||
Expression of ZSCAN10:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ZSCAN10 | 84891 | 1553875_s_at | -0.0236 | 0.9458 | |
| GSE26886 | ZSCAN10 | 84891 | 1553875_s_at | -0.0737 | 0.6237 | |
| GSE45670 | ZSCAN10 | 84891 | 1553875_s_at | 0.0851 | 0.3184 | |
| GSE53622 | ZSCAN10 | 84891 | 32699 | 0.2324 | 0.0841 | |
| GSE53624 | ZSCAN10 | 84891 | 32699 | 0.2362 | 0.0054 | |
| GSE63941 | ZSCAN10 | 84891 | 1553875_s_at | 0.3302 | 0.0458 | |
| GSE77861 | ZSCAN10 | 84891 | 1553875_s_at | 0.0218 | 0.8748 | |
| TCGA | ZSCAN10 | 84891 | RNAseq | 2.1748 | 0.0457 |
Upregulated datasets: 1; Downregulated datasets: 0.
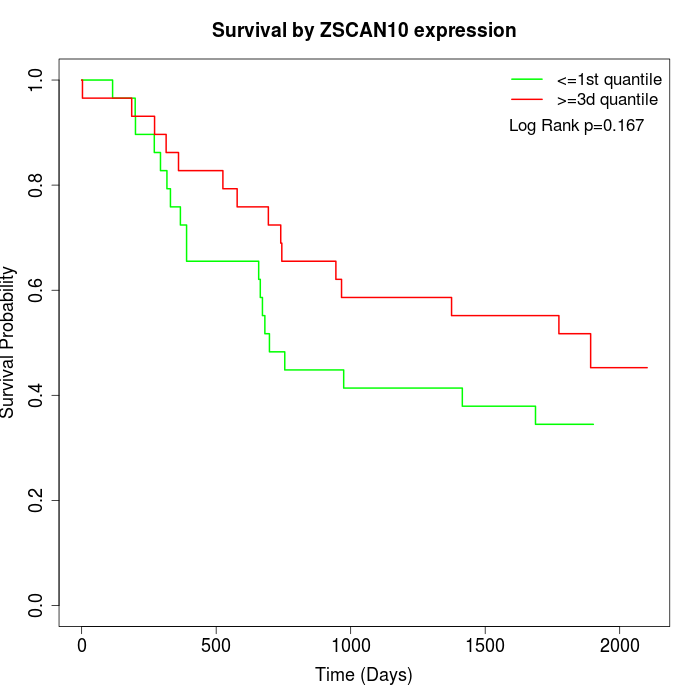
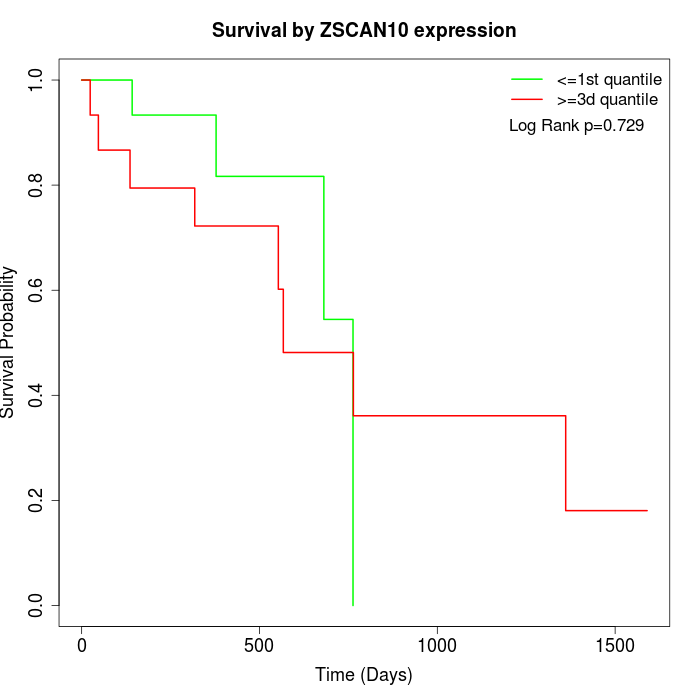
Survival by ZSCAN10 expression:
Note: Click image to view full size file.
Copy number change of ZSCAN10:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ZSCAN10 | 84891 | 5 | 5 | 20 | |
| GSE20123 | ZSCAN10 | 84891 | 5 | 4 | 21 | |
| GSE43470 | ZSCAN10 | 84891 | 3 | 8 | 32 | |
| GSE46452 | ZSCAN10 | 84891 | 38 | 1 | 20 | |
| GSE47630 | ZSCAN10 | 84891 | 13 | 6 | 21 | |
| GSE54993 | ZSCAN10 | 84891 | 3 | 5 | 62 | |
| GSE54994 | ZSCAN10 | 84891 | 5 | 9 | 39 | |
| GSE60625 | ZSCAN10 | 84891 | 4 | 0 | 7 | |
| GSE74703 | ZSCAN10 | 84891 | 3 | 6 | 27 | |
| GSE74704 | ZSCAN10 | 84891 | 3 | 2 | 15 | |
| TCGA | ZSCAN10 | 84891 | 19 | 13 | 64 |
Total number of gains: 101; Total number of losses: 59; Total Number of normals: 328.
Somatic mutations of ZSCAN10:
Generating mutation plots.
Highly correlated genes for ZSCAN10:
Showing top 20/152 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ZSCAN10 | AATK | 0.705129 | 3 | 0 | 3 |
| ZSCAN10 | PTGIR | 0.695366 | 4 | 0 | 4 |
| ZSCAN10 | DSCAM | 0.694601 | 3 | 0 | 3 |
| ZSCAN10 | BTNL3 | 0.687974 | 3 | 0 | 3 |
| ZSCAN10 | IGDCC3 | 0.67991 | 4 | 0 | 4 |
| ZSCAN10 | MYOG | 0.679711 | 3 | 0 | 3 |
| ZSCAN10 | SLITRK2 | 0.678122 | 3 | 0 | 3 |
| ZSCAN10 | FKBP8 | 0.669006 | 3 | 0 | 3 |
| ZSCAN10 | AMHR2 | 0.659259 | 3 | 0 | 3 |
| ZSCAN10 | GRIK5 | 0.656078 | 4 | 0 | 4 |
| ZSCAN10 | TNFRSF4 | 0.656055 | 3 | 0 | 3 |
| ZSCAN10 | MYL10 | 0.652746 | 3 | 0 | 3 |
| ZSCAN10 | AK8 | 0.652083 | 3 | 0 | 3 |
| ZSCAN10 | TM4SF5 | 0.643344 | 4 | 0 | 3 |
| ZSCAN10 | TMEM145 | 0.639962 | 4 | 0 | 3 |
| ZSCAN10 | MESP1 | 0.639006 | 3 | 0 | 3 |
| ZSCAN10 | SSUH2 | 0.638988 | 3 | 0 | 3 |
| ZSCAN10 | EPX | 0.638823 | 3 | 0 | 3 |
| ZSCAN10 | DPF1 | 0.636494 | 3 | 0 | 3 |
| ZSCAN10 | GSX1 | 0.635924 | 4 | 0 | 3 |
For details and further investigation, click here