| Full name: Src homology 2 domain containing transforming protein D | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 19p13.3 | ||
| Entrez ID: 56961 | HGNC ID: HGNC:30633 | Ensembl Gene: ENSG00000105251 | OMIM ID: 610481 |
| Drug and gene relationship at DGIdb | |||
Expression of SHD:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SHD | 56961 | 227845_s_at | -0.0370 | 0.9165 | |
| GSE26886 | SHD | 56961 | 227845_s_at | 0.0168 | 0.9268 | |
| GSE45670 | SHD | 56961 | 227845_s_at | 0.0087 | 0.9448 | |
| GSE53622 | SHD | 56961 | 8913 | 0.1948 | 0.1470 | |
| GSE53624 | SHD | 56961 | 8913 | 0.5623 | 0.0000 | |
| GSE63941 | SHD | 56961 | 227845_s_at | 0.1529 | 0.3563 | |
| GSE77861 | SHD | 56961 | 227845_s_at | -0.1227 | 0.1869 | |
| GSE97050 | SHD | 56961 | A_23_P142255 | 0.1120 | 0.5945 | |
| TCGA | SHD | 56961 | RNAseq | 2.3286 | 0.0484 |
Upregulated datasets: 1; Downregulated datasets: 0.
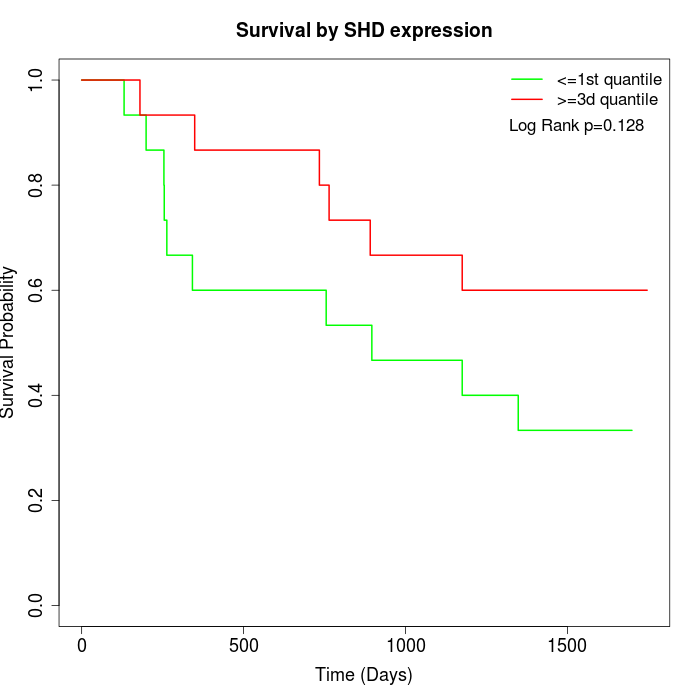
Survival by SHD expression:
Note: Click image to view full size file.
Copy number change of SHD:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SHD | 56961 | 5 | 3 | 22 | |
| GSE20123 | SHD | 56961 | 4 | 2 | 24 | |
| GSE43470 | SHD | 56961 | 1 | 8 | 34 | |
| GSE46452 | SHD | 56961 | 47 | 1 | 11 | |
| GSE47630 | SHD | 56961 | 5 | 7 | 28 | |
| GSE54993 | SHD | 56961 | 16 | 3 | 51 | |
| GSE54994 | SHD | 56961 | 6 | 16 | 31 | |
| GSE60625 | SHD | 56961 | 9 | 0 | 2 | |
| GSE74703 | SHD | 56961 | 1 | 5 | 30 | |
| GSE74704 | SHD | 56961 | 1 | 2 | 17 | |
| TCGA | SHD | 56961 | 11 | 18 | 67 |
Total number of gains: 106; Total number of losses: 65; Total Number of normals: 317.
Somatic mutations of SHD:
Generating mutation plots.
Highly correlated genes for SHD:
Showing top 20/72 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SHD | MYLK2 | 0.769718 | 3 | 0 | 3 |
| SHD | GUCY2C | 0.754644 | 3 | 0 | 3 |
| SHD | ASRGL1 | 0.752633 | 3 | 0 | 3 |
| SHD | CACNG8 | 0.741916 | 3 | 0 | 3 |
| SHD | UFSP1 | 0.736157 | 3 | 0 | 3 |
| SHD | CLEC4G | 0.729982 | 3 | 0 | 3 |
| SHD | GPR78 | 0.719492 | 4 | 0 | 3 |
| SHD | C19orf73 | 0.701828 | 4 | 0 | 3 |
| SHD | SLC25A48 | 0.700151 | 3 | 0 | 3 |
| SHD | SLC25A47 | 0.699633 | 3 | 0 | 3 |
| SHD | KCNH3 | 0.691794 | 3 | 0 | 3 |
| SHD | RASSF7 | 0.688873 | 3 | 0 | 3 |
| SHD | FAM171A2 | 0.687829 | 3 | 0 | 3 |
| SHD | KIF6 | 0.681006 | 3 | 0 | 3 |
| SHD | PRRG3 | 0.680039 | 3 | 0 | 3 |
| SHD | ITIH3 | 0.67853 | 4 | 0 | 3 |
| SHD | KCNK9 | 0.676829 | 3 | 0 | 3 |
| SHD | DHRS7C | 0.676507 | 3 | 0 | 3 |
| SHD | HRC | 0.6734 | 3 | 0 | 3 |
| SHD | CEBPE | 0.673288 | 3 | 0 | 3 |
For details and further investigation, click here