| Full name: C-type lectin domain family 4 member G | Alias Symbol: UNQ431|LSECtin | ||
| Type: protein-coding gene | Cytoband: 19p13.2 | ||
| Entrez ID: 339390 | HGNC ID: HGNC:24591 | Ensembl Gene: ENSG00000182566 | OMIM ID: 616256 |
| Drug and gene relationship at DGIdb | |||
Expression of CLEC4G:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CLEC4G | 339390 | 1559065_a_at | -0.2070 | 0.5476 | |
| GSE26886 | CLEC4G | 339390 | 1559065_a_at | 0.0492 | 0.7073 | |
| GSE45670 | CLEC4G | 339390 | 1559065_a_at | -0.1275 | 0.1631 | |
| GSE53622 | CLEC4G | 339390 | 51456 | -0.2290 | 0.0014 | |
| GSE53624 | CLEC4G | 339390 | 51456 | -0.3134 | 0.0006 | |
| GSE63941 | CLEC4G | 339390 | 1559065_a_at | -0.2503 | 0.1008 | |
| GSE77861 | CLEC4G | 339390 | 1559065_a_at | -0.0677 | 0.5066 | |
| GSE97050 | CLEC4G | 339390 | A_23_P153390 | -0.0680 | 0.7983 | |
| TCGA | CLEC4G | 339390 | RNAseq | -0.6374 | 0.4719 |
Upregulated datasets: 0; Downregulated datasets: 0.
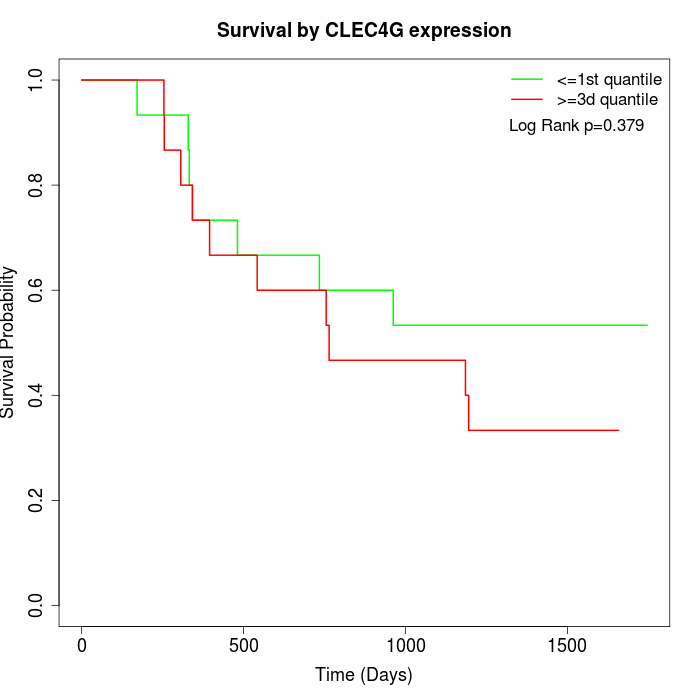
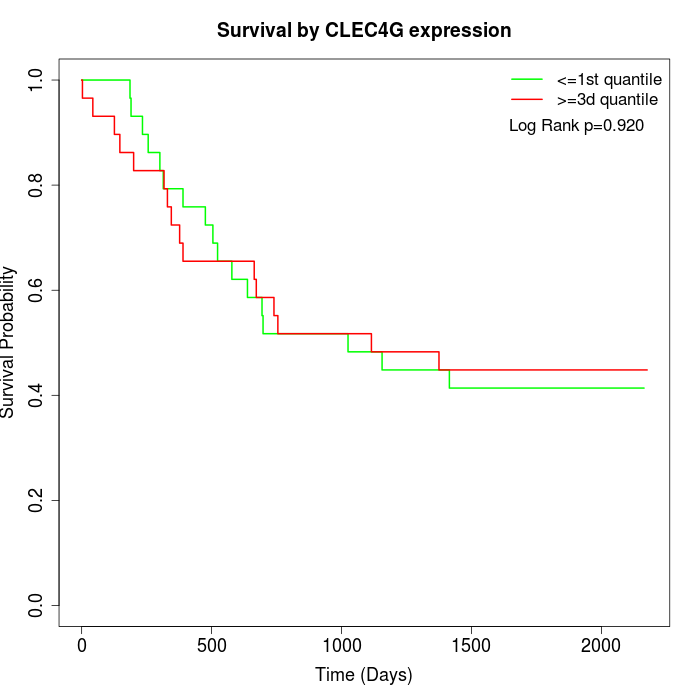
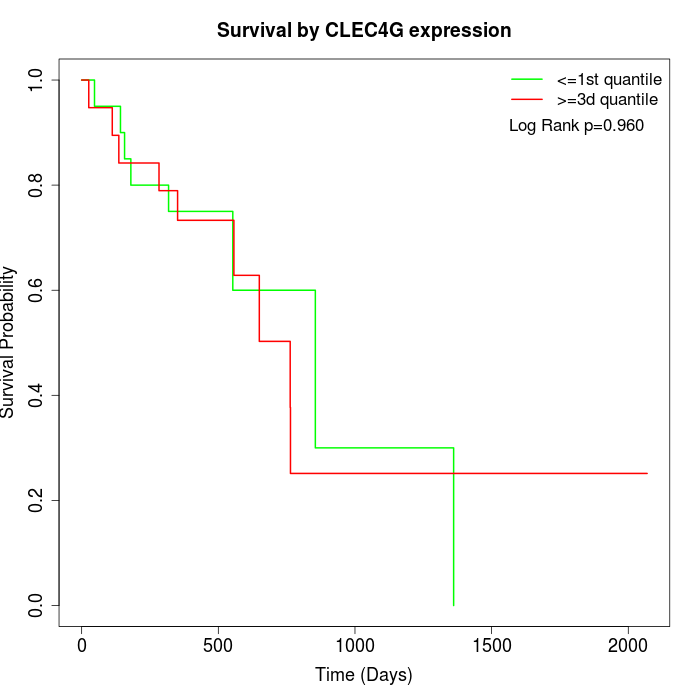
Survival by CLEC4G expression:
Note: Click image to view full size file.
Copy number change of CLEC4G:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CLEC4G | 339390 | 4 | 3 | 23 | |
| GSE20123 | CLEC4G | 339390 | 3 | 2 | 25 | |
| GSE43470 | CLEC4G | 339390 | 2 | 6 | 35 | |
| GSE46452 | CLEC4G | 339390 | 47 | 1 | 11 | |
| GSE47630 | CLEC4G | 339390 | 5 | 7 | 28 | |
| GSE54993 | CLEC4G | 339390 | 16 | 3 | 51 | |
| GSE54994 | CLEC4G | 339390 | 6 | 13 | 34 | |
| GSE60625 | CLEC4G | 339390 | 9 | 0 | 2 | |
| GSE74703 | CLEC4G | 339390 | 2 | 4 | 30 | |
| GSE74704 | CLEC4G | 339390 | 0 | 2 | 18 | |
| TCGA | CLEC4G | 339390 | 9 | 20 | 67 |
Total number of gains: 103; Total number of losses: 61; Total Number of normals: 324.
Somatic mutations of CLEC4G:
Generating mutation plots.
Highly correlated genes for CLEC4G:
Showing top 20/82 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CLEC4G | FGF17 | 0.77268 | 3 | 0 | 3 |
| CLEC4G | CACNA1I | 0.752334 | 3 | 0 | 3 |
| CLEC4G | FRMD1 | 0.743129 | 3 | 0 | 3 |
| CLEC4G | KRTAP19-1 | 0.739014 | 3 | 0 | 3 |
| CLEC4G | SHD | 0.729982 | 3 | 0 | 3 |
| CLEC4G | SNAI1 | 0.707101 | 3 | 0 | 3 |
| CLEC4G | NLRP6 | 0.698873 | 3 | 0 | 3 |
| CLEC4G | SYCE3 | 0.697784 | 3 | 0 | 3 |
| CLEC4G | KANK3 | 0.692768 | 4 | 0 | 4 |
| CLEC4G | GPR78 | 0.682528 | 3 | 0 | 3 |
| CLEC4G | CACNG8 | 0.681866 | 4 | 0 | 4 |
| CLEC4G | MYH13 | 0.671695 | 3 | 0 | 3 |
| CLEC4G | PRSS54 | 0.671035 | 3 | 0 | 3 |
| CLEC4G | C20orf85 | 0.670082 | 4 | 0 | 3 |
| CLEC4G | C8G | 0.667864 | 4 | 0 | 3 |
| CLEC4G | NOBOX | 0.665956 | 4 | 0 | 3 |
| CLEC4G | PALM3 | 0.665419 | 4 | 0 | 3 |
| CLEC4G | TOR2A | 0.658975 | 4 | 0 | 4 |
| CLEC4G | GLTPD2 | 0.655919 | 3 | 0 | 3 |
| CLEC4G | OR4D2 | 0.651212 | 4 | 0 | 3 |
For details and further investigation, click here