| Full name: trace amine associated receptor 2 (gene/pseudogene) | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 6q23.2 | ||
| Entrez ID: 9287 | HGNC ID: HGNC:4514 | Ensembl Gene: ENSG00000146378 | OMIM ID: 604849 |
| Drug and gene relationship at DGIdb | |||
Expression of TAAR2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TAAR2 | 9287 | 221394_at | -0.1296 | 0.5186 | |
| GSE20347 | TAAR2 | 9287 | 221394_at | -0.0137 | 0.8704 | |
| GSE23400 | TAAR2 | 9287 | 221394_at | -0.2380 | 0.0000 | |
| GSE26886 | TAAR2 | 9287 | 221394_at | 0.0052 | 0.9669 | |
| GSE29001 | TAAR2 | 9287 | 221394_at | -0.2517 | 0.1430 | |
| GSE38129 | TAAR2 | 9287 | 221394_at | -0.1417 | 0.0593 | |
| GSE45670 | TAAR2 | 9287 | 221394_at | 0.0616 | 0.5608 | |
| GSE53622 | TAAR2 | 9287 | 80857 | 0.0577 | 0.5326 | |
| GSE53624 | TAAR2 | 9287 | 80857 | 0.2440 | 0.0001 | |
| GSE63941 | TAAR2 | 9287 | 221394_at | -0.0357 | 0.8248 | |
| GSE77861 | TAAR2 | 9287 | 221394_at | -0.0513 | 0.4758 |
Upregulated datasets: 0; Downregulated datasets: 0.
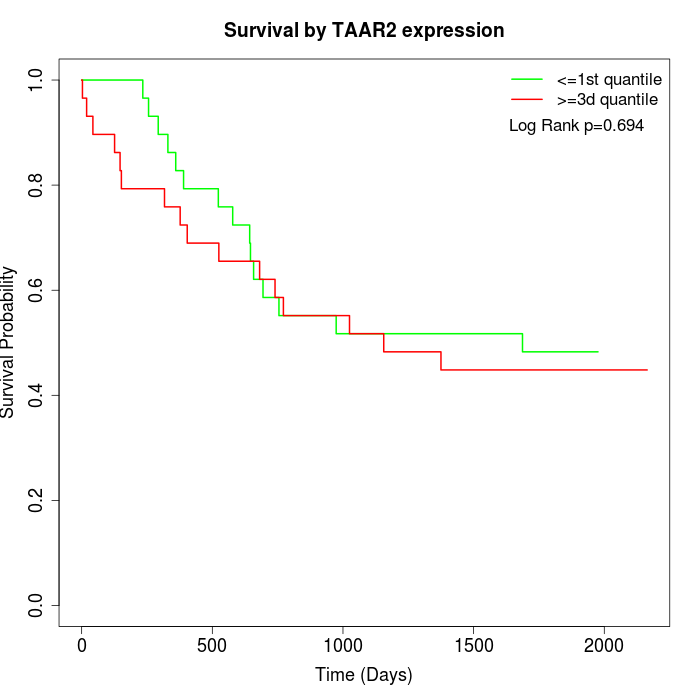
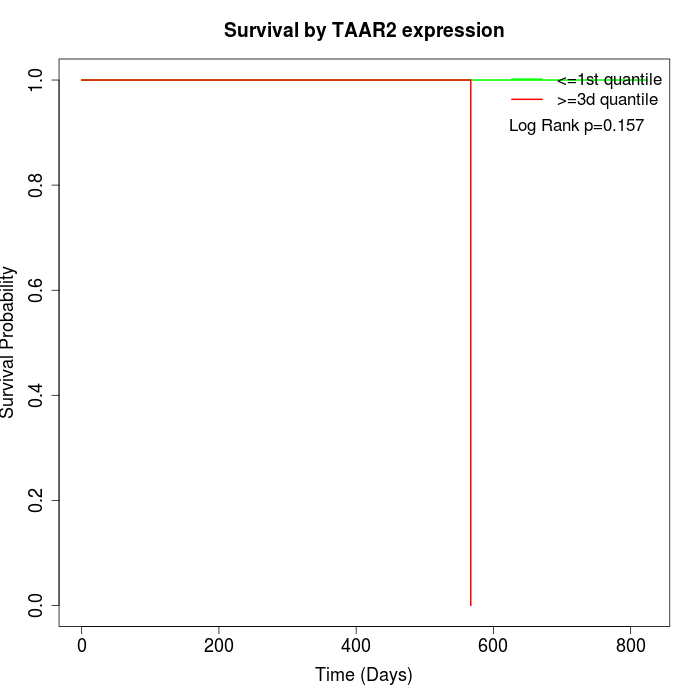
Survival by TAAR2 expression:
Note: Click image to view full size file.
Copy number change of TAAR2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TAAR2 | 9287 | 3 | 4 | 23 | |
| GSE20123 | TAAR2 | 9287 | 3 | 3 | 24 | |
| GSE43470 | TAAR2 | 9287 | 5 | 0 | 38 | |
| GSE46452 | TAAR2 | 9287 | 2 | 11 | 46 | |
| GSE47630 | TAAR2 | 9287 | 9 | 4 | 27 | |
| GSE54993 | TAAR2 | 9287 | 3 | 2 | 65 | |
| GSE54994 | TAAR2 | 9287 | 9 | 7 | 37 | |
| GSE60625 | TAAR2 | 9287 | 0 | 1 | 10 | |
| GSE74703 | TAAR2 | 9287 | 4 | 0 | 32 | |
| GSE74704 | TAAR2 | 9287 | 1 | 1 | 18 | |
| TCGA | TAAR2 | 9287 | 11 | 18 | 67 |
Total number of gains: 50; Total number of losses: 51; Total Number of normals: 387.
Somatic mutations of TAAR2:
Generating mutation plots.
Highly correlated genes for TAAR2:
Showing top 20/891 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TAAR2 | CAMK4 | 0.776203 | 3 | 0 | 3 |
| TAAR2 | PSG9 | 0.739221 | 4 | 0 | 4 |
| TAAR2 | TSHB | 0.724165 | 4 | 0 | 4 |
| TAAR2 | ART1 | 0.724025 | 4 | 0 | 3 |
| TAAR2 | HHLA1 | 0.719096 | 4 | 0 | 4 |
| TAAR2 | UGT2A3 | 0.717455 | 3 | 0 | 3 |
| TAAR2 | EPX | 0.713948 | 4 | 0 | 4 |
| TAAR2 | SLC17A3 | 0.709086 | 3 | 0 | 3 |
| TAAR2 | ATP4A | 0.707534 | 3 | 0 | 3 |
| TAAR2 | ADM2 | 0.704231 | 4 | 0 | 4 |
| TAAR2 | IGH | 0.703554 | 3 | 0 | 3 |
| TAAR2 | GRPR | 0.701955 | 4 | 0 | 4 |
| TAAR2 | PAX3 | 0.698711 | 5 | 0 | 4 |
| TAAR2 | RHO | 0.6986 | 3 | 0 | 3 |
| TAAR2 | SPX | 0.697257 | 5 | 0 | 4 |
| TAAR2 | DRD5 | 0.693899 | 4 | 0 | 4 |
| TAAR2 | LINC01260 | 0.692649 | 3 | 0 | 3 |
| TAAR2 | OR7C1 | 0.691939 | 4 | 0 | 4 |
| TAAR2 | NEURL1 | 0.688649 | 4 | 0 | 4 |
| TAAR2 | IFNA8 | 0.685997 | 3 | 0 | 3 |
For details and further investigation, click here