| Full name: eosinophil peroxidase | Alias Symbol: EPO|EPP|EPX-PEN | ||
| Type: protein-coding gene | Cytoband: 17q22 | ||
| Entrez ID: 8288 | HGNC ID: HGNC:3423 | Ensembl Gene: ENSG00000121053 | OMIM ID: 131399 |
| Drug and gene relationship at DGIdb | |||
Expression of EPX:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | EPX | 8288 | 214627_at | 0.0284 | 0.9350 | |
| GSE20347 | EPX | 8288 | 214627_at | 0.0306 | 0.7603 | |
| GSE23400 | EPX | 8288 | 214627_at | -0.1124 | 0.0145 | |
| GSE26886 | EPX | 8288 | 214627_at | 0.1234 | 0.2385 | |
| GSE29001 | EPX | 8288 | 214627_at | -0.0589 | 0.7037 | |
| GSE38129 | EPX | 8288 | 214627_at | -0.0569 | 0.5775 | |
| GSE45670 | EPX | 8288 | 214627_at | 0.1394 | 0.1207 | |
| GSE53622 | EPX | 8288 | 143811 | -0.3138 | 0.0144 | |
| GSE53624 | EPX | 8288 | 143811 | -0.2572 | 0.0054 | |
| GSE63941 | EPX | 8288 | 214627_at | 0.3425 | 0.0381 | |
| GSE77861 | EPX | 8288 | 214627_at | -0.0436 | 0.7646 | |
| TCGA | EPX | 8288 | RNAseq | -0.2939 | 0.5572 |
Upregulated datasets: 0; Downregulated datasets: 0.
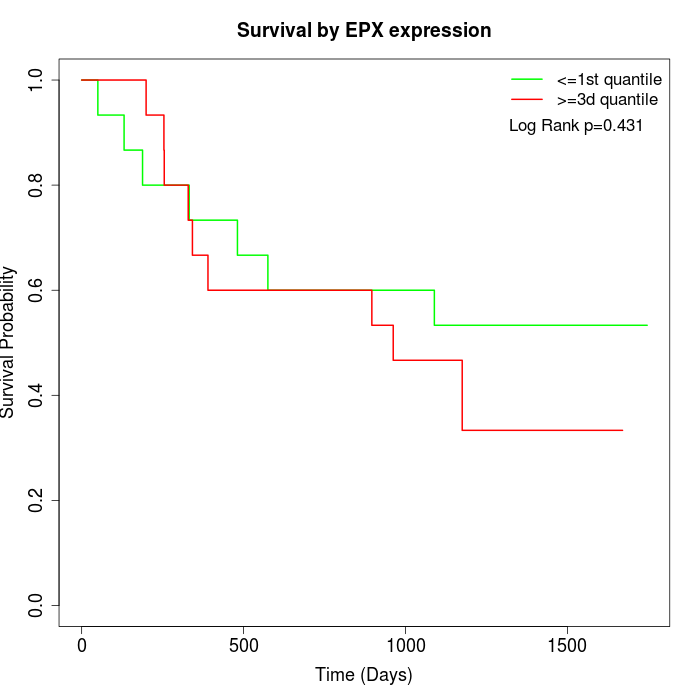
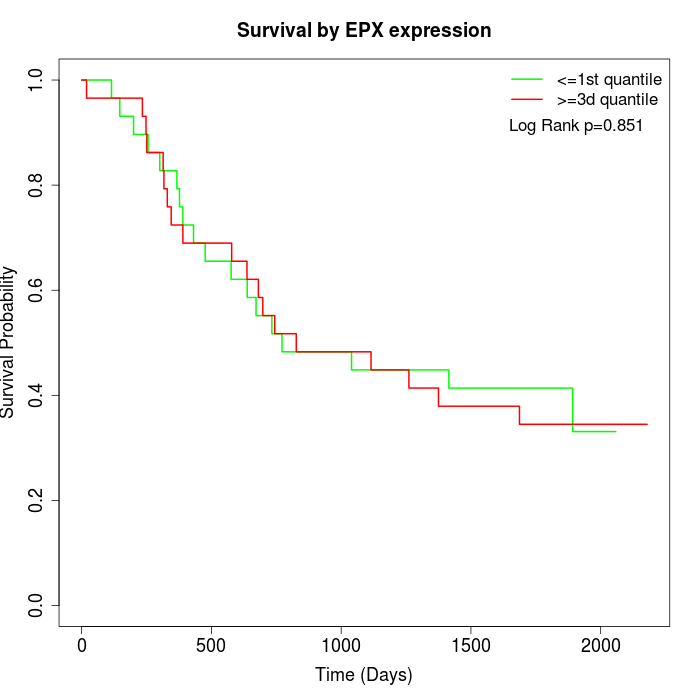
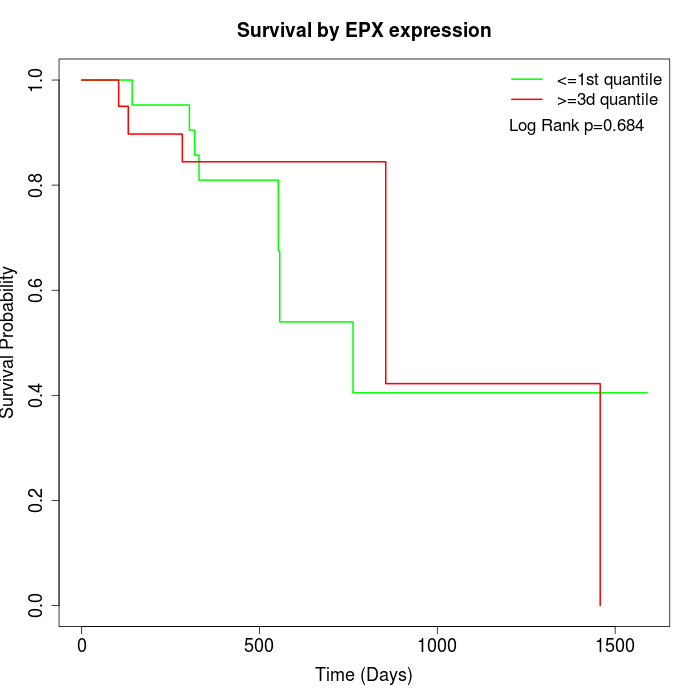
Survival by EPX expression:
Note: Click image to view full size file.
Copy number change of EPX:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | EPX | 8288 | 5 | 1 | 24 | |
| GSE20123 | EPX | 8288 | 5 | 1 | 24 | |
| GSE43470 | EPX | 8288 | 4 | 0 | 39 | |
| GSE46452 | EPX | 8288 | 32 | 0 | 27 | |
| GSE47630 | EPX | 8288 | 7 | 1 | 32 | |
| GSE54993 | EPX | 8288 | 2 | 5 | 63 | |
| GSE54994 | EPX | 8288 | 8 | 6 | 39 | |
| GSE60625 | EPX | 8288 | 4 | 0 | 7 | |
| GSE74703 | EPX | 8288 | 4 | 0 | 32 | |
| GSE74704 | EPX | 8288 | 4 | 1 | 15 | |
| TCGA | EPX | 8288 | 26 | 8 | 62 |
Total number of gains: 101; Total number of losses: 23; Total Number of normals: 364.
Somatic mutations of EPX:
Generating mutation plots.
Highly correlated genes for EPX:
Showing top 20/1257 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| EPX | LINC00642 | 0.757056 | 3 | 0 | 3 |
| EPX | ADM2 | 0.744769 | 6 | 0 | 6 |
| EPX | G6PC2 | 0.734901 | 4 | 0 | 4 |
| EPX | GAD2 | 0.728179 | 4 | 0 | 4 |
| EPX | P2RX6 | 0.724531 | 5 | 0 | 5 |
| EPX | C6 | 0.715567 | 4 | 0 | 4 |
| EPX | TAAR2 | 0.713948 | 4 | 0 | 4 |
| EPX | MAPK15 | 0.713139 | 3 | 0 | 3 |
| EPX | GSX1 | 0.711648 | 3 | 0 | 3 |
| EPX | CELF3 | 0.70787 | 7 | 0 | 6 |
| EPX | USP29 | 0.706186 | 4 | 0 | 4 |
| EPX | ADCYAP1 | 0.705013 | 3 | 0 | 3 |
| EPX | NPTXR | 0.704809 | 4 | 0 | 4 |
| EPX | EDA2R | 0.70183 | 5 | 0 | 5 |
| EPX | NTNG1 | 0.697242 | 4 | 0 | 4 |
| EPX | ATF7 | 0.695447 | 4 | 0 | 4 |
| EPX | SLC6A5 | 0.695123 | 4 | 0 | 4 |
| EPX | JAK3 | 0.694494 | 6 | 0 | 6 |
| EPX | GIGYF1 | 0.694376 | 3 | 0 | 3 |
| EPX | LINC01339 | 0.692248 | 3 | 0 | 3 |
For details and further investigation, click here