| Full name: taste 2 receptor member 50 | Alias Symbol: T2R51 | ||
| Type: protein-coding gene | Cytoband: 12p13.2 | ||
| Entrez ID: 259296 | HGNC ID: HGNC:18882 | Ensembl Gene: ENSG00000212126 | OMIM ID: 609627 |
| Drug and gene relationship at DGIdb | |||
Expression of TAS2R50:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TAS2R50 | 259296 | 1553561_at | -0.0025 | 0.9941 | |
| GSE26886 | TAS2R50 | 259296 | 1553561_at | -0.1078 | 0.3145 | |
| GSE45670 | TAS2R50 | 259296 | 1553561_at | 0.0158 | 0.8496 | |
| GSE53622 | TAS2R50 | 259296 | 117597 | 0.8358 | 0.0000 | |
| GSE53624 | TAS2R50 | 259296 | 117597 | 0.4878 | 0.0036 | |
| GSE63941 | TAS2R50 | 259296 | 1553561_at | 0.2625 | 0.1640 | |
| GSE77861 | TAS2R50 | 259296 | 1553561_at | -0.0971 | 0.2959 | |
| GSE97050 | TAS2R50 | 259296 | A_23_P347029 | 0.2865 | 0.2513 | |
| SRP133303 | TAS2R50 | 259296 | RNAseq | 0.4858 | 0.1284 | |
| TCGA | TAS2R50 | 259296 | RNAseq | -0.8941 | 0.1933 |
Upregulated datasets: 0; Downregulated datasets: 0.
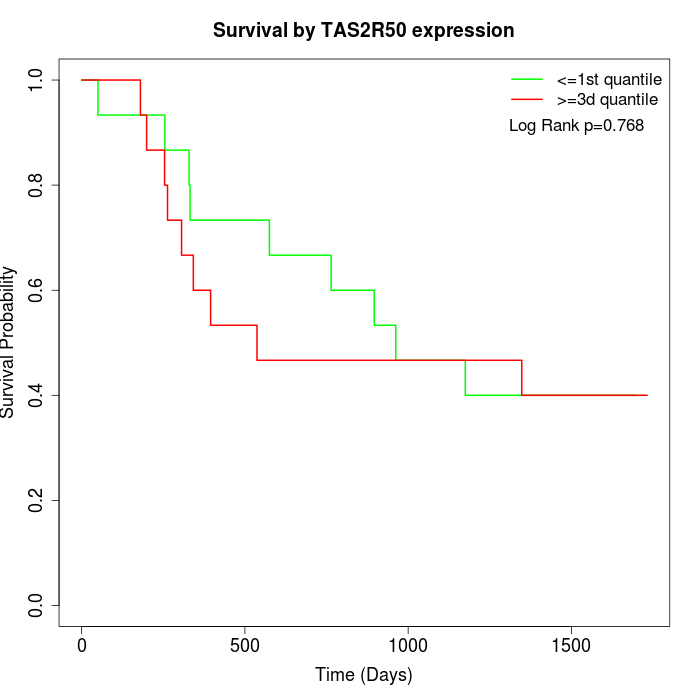
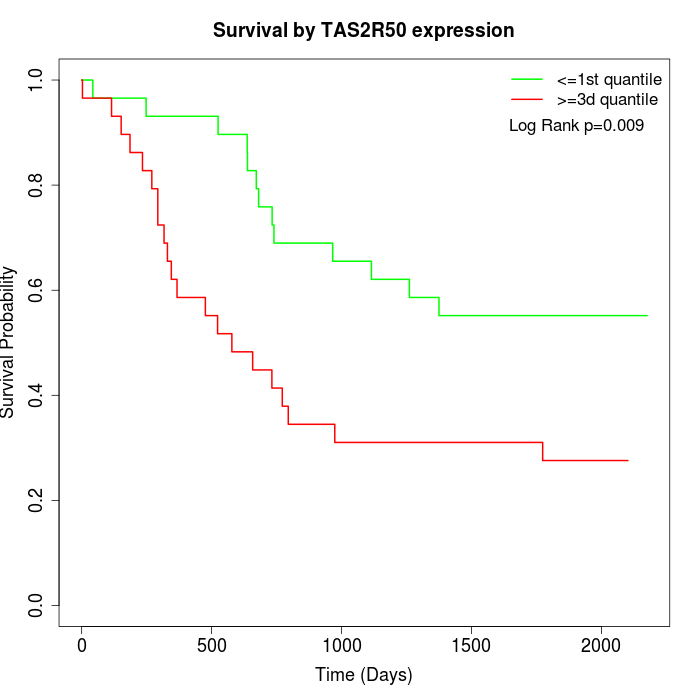
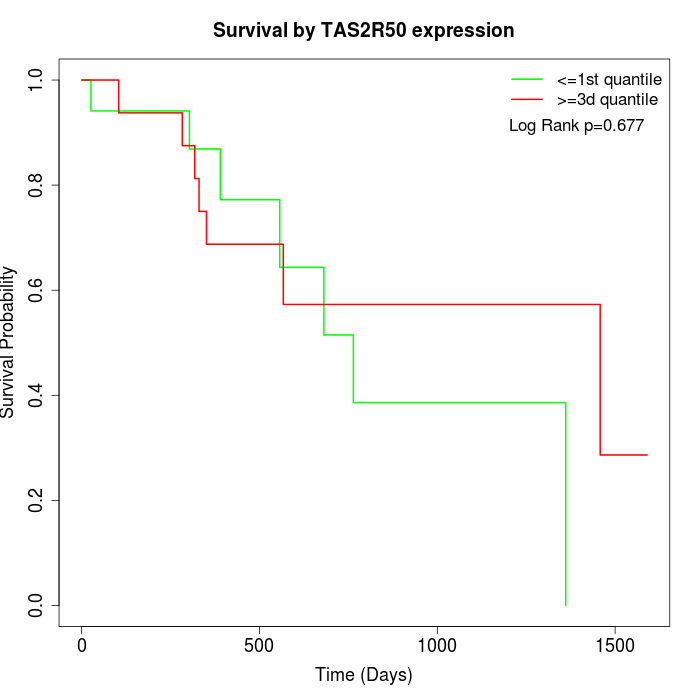
Survival by TAS2R50 expression:
Note: Click image to view full size file.
Copy number change of TAS2R50:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TAS2R50 | 259296 | 6 | 2 | 22 | |
| GSE20123 | TAS2R50 | 259296 | 6 | 2 | 22 | |
| GSE43470 | TAS2R50 | 259296 | 9 | 3 | 31 | |
| GSE46452 | TAS2R50 | 259296 | 10 | 1 | 48 | |
| GSE47630 | TAS2R50 | 259296 | 14 | 2 | 24 | |
| GSE54993 | TAS2R50 | 259296 | 1 | 10 | 59 | |
| GSE54994 | TAS2R50 | 259296 | 9 | 2 | 42 | |
| GSE60625 | TAS2R50 | 259296 | 0 | 1 | 10 | |
| GSE74703 | TAS2R50 | 259296 | 9 | 2 | 25 | |
| GSE74704 | TAS2R50 | 259296 | 4 | 1 | 15 | |
| TCGA | TAS2R50 | 259296 | 37 | 8 | 51 |
Total number of gains: 105; Total number of losses: 34; Total Number of normals: 349.
Somatic mutations of TAS2R50:
Generating mutation plots.
Highly correlated genes for TAS2R50:
Showing top 20/36 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TAS2R50 | TAS2R43 | 0.879738 | 3 | 0 | 3 |
| TAS2R50 | TAS2R19 | 0.815264 | 4 | 0 | 4 |
| TAS2R50 | TAS2R20 | 0.81069 | 3 | 0 | 3 |
| TAS2R50 | TAS2R46 | 0.81069 | 3 | 0 | 3 |
| TAS2R50 | C19orf54 | 0.717112 | 3 | 0 | 3 |
| TAS2R50 | ACAD11 | 0.711354 | 3 | 0 | 3 |
| TAS2R50 | ZNF35 | 0.706849 | 3 | 0 | 3 |
| TAS2R50 | SPDYE1 | 0.706249 | 3 | 0 | 3 |
| TAS2R50 | ZNF480 | 0.696257 | 3 | 0 | 3 |
| TAS2R50 | RBBP5 | 0.691387 | 3 | 0 | 3 |
| TAS2R50 | ARL17A | 0.68472 | 3 | 0 | 3 |
| TAS2R50 | ALG11 | 0.684418 | 3 | 0 | 3 |
| TAS2R50 | ZNRD1 | 0.683917 | 3 | 0 | 3 |
| TAS2R50 | ZNF71 | 0.675054 | 3 | 0 | 3 |
| TAS2R50 | SPDYE5 | 0.66935 | 3 | 0 | 3 |
| TAS2R50 | TMEM18 | 0.668942 | 3 | 0 | 3 |
| TAS2R50 | BTN1A1 | 0.662657 | 3 | 0 | 3 |
| TAS2R50 | ZNF248 | 0.660877 | 3 | 0 | 3 |
| TAS2R50 | CEP57 | 0.658554 | 3 | 0 | 3 |
| TAS2R50 | DHODH | 0.653119 | 3 | 0 | 3 |
For details and further investigation, click here