| Full name: centrosomal protein 57 | Alias Symbol: Translokin|TSP57|KIAA0092 | ||
| Type: protein-coding gene | Cytoband: 11q21 | ||
| Entrez ID: 9702 | HGNC ID: HGNC:30794 | Ensembl Gene: ENSG00000166037 | OMIM ID: 607951 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of CEP57:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CEP57 | 9702 | 203494_s_at | -0.3985 | 0.1165 | |
| GSE20347 | CEP57 | 9702 | 203494_s_at | -0.1092 | 0.6120 | |
| GSE23400 | CEP57 | 9702 | 203494_s_at | 0.2207 | 0.0191 | |
| GSE26886 | CEP57 | 9702 | 203494_s_at | 0.4053 | 0.1409 | |
| GSE29001 | CEP57 | 9702 | 203494_s_at | -0.0955 | 0.7685 | |
| GSE38129 | CEP57 | 9702 | 203494_s_at | -0.1389 | 0.3552 | |
| GSE45670 | CEP57 | 9702 | 203494_s_at | -0.3516 | 0.1057 | |
| GSE53622 | CEP57 | 9702 | 10504 | 0.2740 | 0.0005 | |
| GSE53624 | CEP57 | 9702 | 10504 | 0.3054 | 0.0001 | |
| GSE63941 | CEP57 | 9702 | 203492_x_at | 1.0207 | 0.0771 | |
| GSE77861 | CEP57 | 9702 | 203492_x_at | 0.2618 | 0.5108 | |
| GSE97050 | CEP57 | 9702 | A_23_P86822 | 0.0872 | 0.8428 | |
| SRP007169 | CEP57 | 9702 | RNAseq | 0.2795 | 0.5672 | |
| SRP008496 | CEP57 | 9702 | RNAseq | 0.1256 | 0.6203 | |
| SRP064894 | CEP57 | 9702 | RNAseq | -0.0688 | 0.7303 | |
| SRP133303 | CEP57 | 9702 | RNAseq | -0.1633 | 0.2517 | |
| SRP159526 | CEP57 | 9702 | RNAseq | 0.0027 | 0.9929 | |
| SRP193095 | CEP57 | 9702 | RNAseq | -0.4280 | 0.0005 | |
| SRP219564 | CEP57 | 9702 | RNAseq | -0.0475 | 0.8016 | |
| TCGA | CEP57 | 9702 | RNAseq | -0.0267 | 0.6212 |
Upregulated datasets: 0; Downregulated datasets: 0.
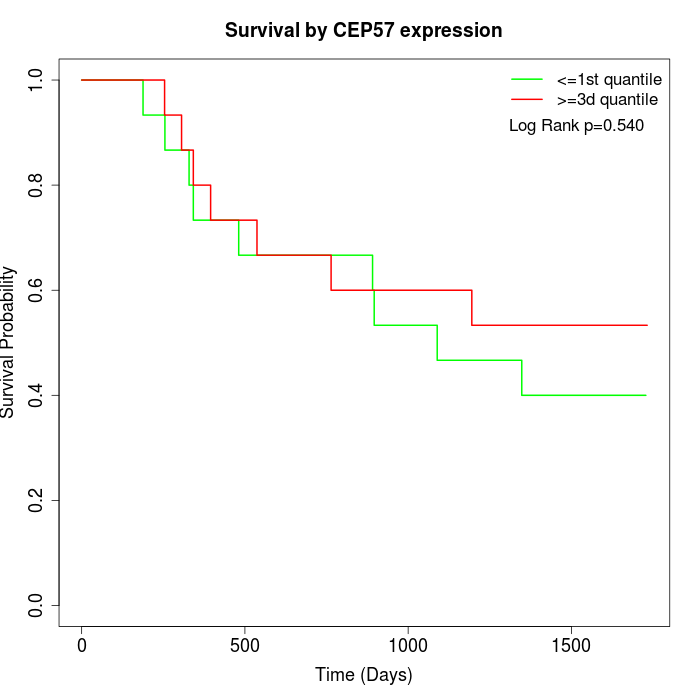
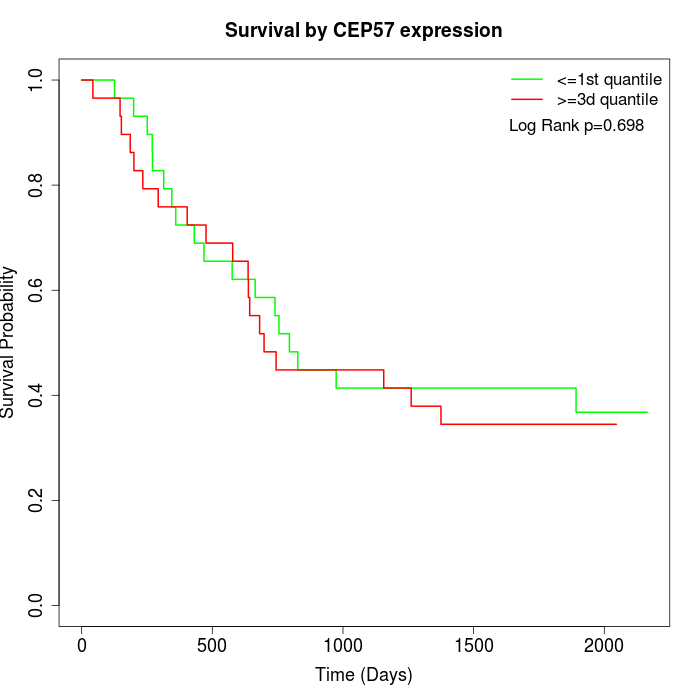
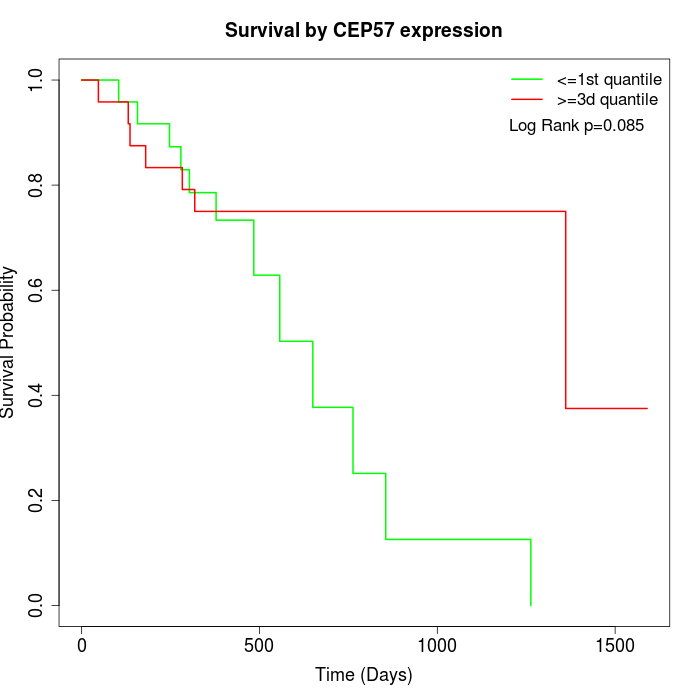
Survival by CEP57 expression:
Note: Click image to view full size file.
Copy number change of CEP57:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CEP57 | 9702 | 3 | 9 | 18 | |
| GSE20123 | CEP57 | 9702 | 3 | 9 | 18 | |
| GSE43470 | CEP57 | 9702 | 2 | 6 | 35 | |
| GSE46452 | CEP57 | 9702 | 5 | 24 | 30 | |
| GSE47630 | CEP57 | 9702 | 2 | 19 | 19 | |
| GSE54993 | CEP57 | 9702 | 9 | 2 | 59 | |
| GSE54994 | CEP57 | 9702 | 6 | 18 | 29 | |
| GSE60625 | CEP57 | 9702 | 0 | 3 | 8 | |
| GSE74703 | CEP57 | 9702 | 1 | 5 | 30 | |
| GSE74704 | CEP57 | 9702 | 3 | 5 | 12 | |
| TCGA | CEP57 | 9702 | 11 | 42 | 43 |
Total number of gains: 45; Total number of losses: 142; Total Number of normals: 301.
Somatic mutations of CEP57:
Generating mutation plots.
Highly correlated genes for CEP57:
Showing top 20/237 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CEP57 | C1orf50 | 0.821652 | 3 | 0 | 3 |
| CEP57 | GAPT | 0.766888 | 3 | 0 | 3 |
| CEP57 | RECK | 0.766349 | 3 | 0 | 3 |
| CEP57 | KLHL15 | 0.759613 | 3 | 0 | 3 |
| CEP57 | RPUSD3 | 0.751379 | 3 | 0 | 3 |
| CEP57 | RAB40AL | 0.744816 | 3 | 0 | 3 |
| CEP57 | CGRRF1 | 0.740922 | 3 | 0 | 3 |
| CEP57 | ZNF548 | 0.735426 | 3 | 0 | 3 |
| CEP57 | HOXA3 | 0.733634 | 3 | 0 | 3 |
| CEP57 | HSF2 | 0.726347 | 3 | 0 | 3 |
| CEP57 | DLD | 0.722498 | 3 | 0 | 3 |
| CEP57 | GMCL1 | 0.715466 | 3 | 0 | 3 |
| CEP57 | MOB3B | 0.711895 | 3 | 0 | 3 |
| CEP57 | PDK3 | 0.707935 | 3 | 0 | 3 |
| CEP57 | CALM1 | 0.700197 | 3 | 0 | 3 |
| CEP57 | TAS2R19 | 0.688751 | 3 | 0 | 3 |
| CEP57 | ANAPC5 | 0.680874 | 3 | 0 | 3 |
| CEP57 | SLC19A2 | 0.680386 | 3 | 0 | 3 |
| CEP57 | PIAS2 | 0.680099 | 4 | 0 | 4 |
| CEP57 | PTPMT1 | 0.676861 | 3 | 0 | 3 |
For details and further investigation, click here