| Full name: transcription elongation regulator 1 | Alias Symbol: CA150|Urn1 | ||
| Type: protein-coding gene | Cytoband: 5q32 | ||
| Entrez ID: 10915 | HGNC ID: HGNC:15630 | Ensembl Gene: ENSG00000113649 | OMIM ID: 605409 |
| Drug and gene relationship at DGIdb | |||
Expression of TCERG1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TCERG1 | 10915 | 202396_at | 0.2978 | 0.4792 | |
| GSE20347 | TCERG1 | 10915 | 202396_at | 0.2181 | 0.0572 | |
| GSE23400 | TCERG1 | 10915 | 202396_at | 0.6287 | 0.0000 | |
| GSE26886 | TCERG1 | 10915 | 202396_at | -0.2513 | 0.1853 | |
| GSE29001 | TCERG1 | 10915 | 202396_at | 0.2399 | 0.3754 | |
| GSE38129 | TCERG1 | 10915 | 202396_at | 0.2826 | 0.0849 | |
| GSE45670 | TCERG1 | 10915 | 202396_at | 0.1055 | 0.4858 | |
| GSE53622 | TCERG1 | 10915 | 121783 | 0.3112 | 0.0000 | |
| GSE53624 | TCERG1 | 10915 | 121783 | 0.2917 | 0.0000 | |
| GSE63941 | TCERG1 | 10915 | 202396_at | 0.6016 | 0.0430 | |
| GSE77861 | TCERG1 | 10915 | 202396_at | 0.3821 | 0.1692 | |
| GSE97050 | TCERG1 | 10915 | A_23_P133365 | 0.2292 | 0.5222 | |
| SRP007169 | TCERG1 | 10915 | RNAseq | 0.5425 | 0.1955 | |
| SRP008496 | TCERG1 | 10915 | RNAseq | 0.4464 | 0.1310 | |
| SRP064894 | TCERG1 | 10915 | RNAseq | -0.0636 | 0.7948 | |
| SRP133303 | TCERG1 | 10915 | RNAseq | 0.2024 | 0.0698 | |
| SRP159526 | TCERG1 | 10915 | RNAseq | -0.1338 | 0.4330 | |
| SRP193095 | TCERG1 | 10915 | RNAseq | 0.1647 | 0.0904 | |
| SRP219564 | TCERG1 | 10915 | RNAseq | 0.2279 | 0.5751 | |
| TCGA | TCERG1 | 10915 | RNAseq | 0.0615 | 0.1867 |
Upregulated datasets: 0; Downregulated datasets: 0.
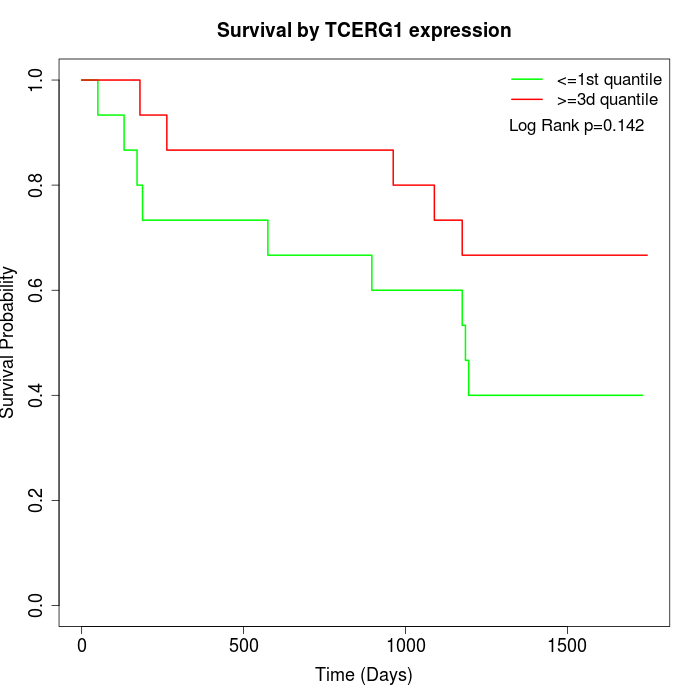
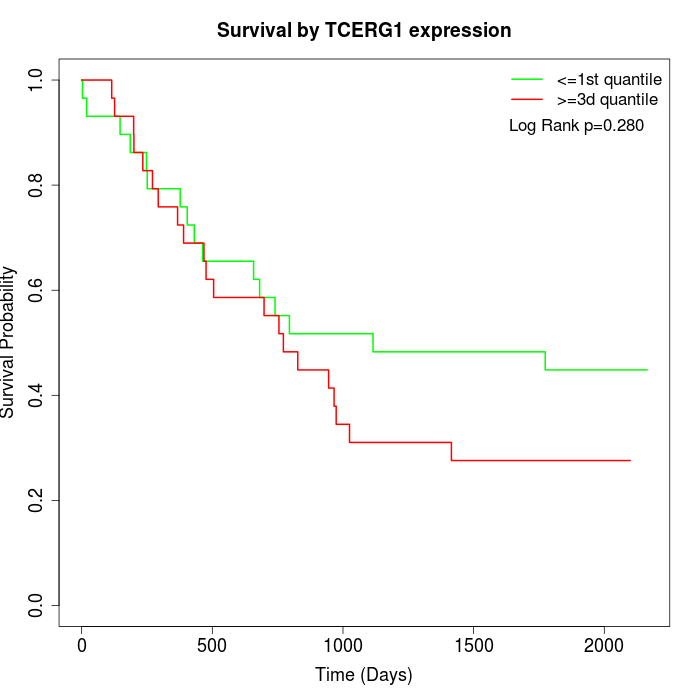
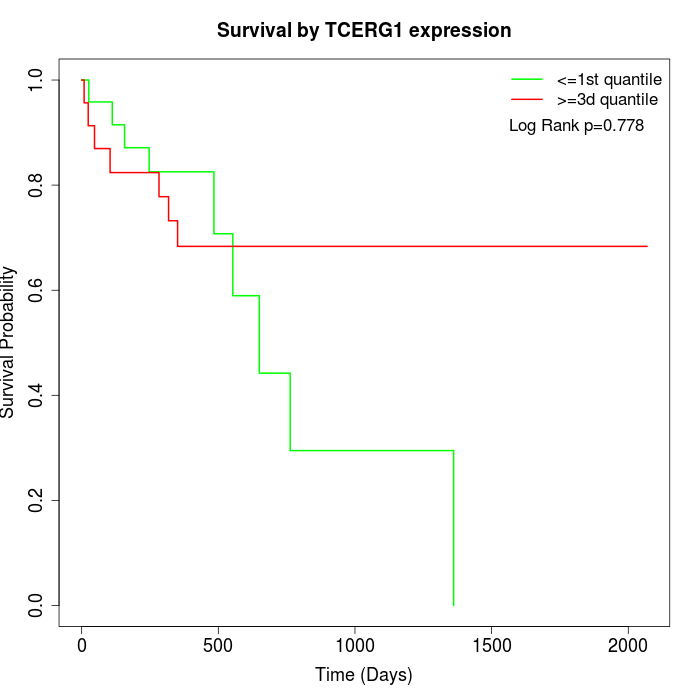
Survival by TCERG1 expression:
Note: Click image to view full size file.
Copy number change of TCERG1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TCERG1 | 10915 | 1 | 12 | 17 | |
| GSE20123 | TCERG1 | 10915 | 1 | 12 | 17 | |
| GSE43470 | TCERG1 | 10915 | 2 | 9 | 32 | |
| GSE46452 | TCERG1 | 10915 | 0 | 27 | 32 | |
| GSE47630 | TCERG1 | 10915 | 0 | 20 | 20 | |
| GSE54993 | TCERG1 | 10915 | 9 | 1 | 60 | |
| GSE54994 | TCERG1 | 10915 | 2 | 14 | 37 | |
| GSE60625 | TCERG1 | 10915 | 0 | 0 | 11 | |
| GSE74703 | TCERG1 | 10915 | 2 | 7 | 27 | |
| GSE74704 | TCERG1 | 10915 | 1 | 7 | 12 | |
| TCGA | TCERG1 | 10915 | 3 | 40 | 53 |
Total number of gains: 21; Total number of losses: 149; Total Number of normals: 318.
Somatic mutations of TCERG1:
Generating mutation plots.
Highly correlated genes for TCERG1:
Showing top 20/788 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TCERG1 | RSPRY1 | 0.784075 | 3 | 0 | 3 |
| TCERG1 | ZGPAT | 0.762631 | 3 | 0 | 3 |
| TCERG1 | ISCA2 | 0.745508 | 3 | 0 | 3 |
| TCERG1 | THBD | 0.741857 | 3 | 0 | 3 |
| TCERG1 | MZT2B | 0.716611 | 4 | 0 | 3 |
| TCERG1 | BTF3L4 | 0.710163 | 3 | 0 | 3 |
| TCERG1 | DIP2A | 0.705625 | 4 | 0 | 3 |
| TCERG1 | SLC45A4 | 0.705168 | 3 | 0 | 3 |
| TCERG1 | TIMM8A | 0.704872 | 3 | 0 | 3 |
| TCERG1 | RLIM | 0.702224 | 4 | 0 | 3 |
| TCERG1 | SLC35E1 | 0.70152 | 4 | 0 | 3 |
| TCERG1 | HSD17B7 | 0.686801 | 3 | 0 | 3 |
| TCERG1 | POLR1B | 0.686577 | 6 | 0 | 5 |
| TCERG1 | ZNF721 | 0.685155 | 4 | 0 | 3 |
| TCERG1 | RELL2 | 0.682105 | 3 | 0 | 3 |
| TCERG1 | SNX27 | 0.680064 | 3 | 0 | 3 |
| TCERG1 | PTPN4 | 0.675954 | 4 | 0 | 3 |
| TCERG1 | HTATSF1 | 0.67397 | 4 | 0 | 4 |
| TCERG1 | MRPL13 | 0.67288 | 6 | 0 | 6 |
| TCERG1 | EIF2A | 0.670952 | 4 | 0 | 3 |
For details and further investigation, click here