| Full name: testis-specific transcript, Y-linked 11 | Alias Symbol: TTY11|NCRNA00134 | ||
| Type: non-coding RNA | Cytoband: Yp11.2 | ||
| Entrez ID: 83866 | HGNC ID: HGNC:18492 | Ensembl Gene: ENSG00000180910 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of TTTY11:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TTTY11 | 83866 | 224174_at | -0.2301 | 0.4577 | |
| GSE26886 | TTTY11 | 83866 | 224174_at | -0.2706 | 0.0482 | |
| GSE45670 | TTTY11 | 83866 | 224174_at | 0.0778 | 0.3783 | |
| GSE53622 | TTTY11 | 83866 | 20463 | 0.3798 | 0.0000 | |
| GSE53624 | TTTY11 | 83866 | 20463 | 0.1618 | 0.1028 | |
| GSE63941 | TTTY11 | 83866 | 224174_at | 0.1852 | 0.3207 | |
| GSE77861 | TTTY11 | 83866 | 224174_at | -0.1685 | 0.0455 |
Upregulated datasets: 0; Downregulated datasets: 0.
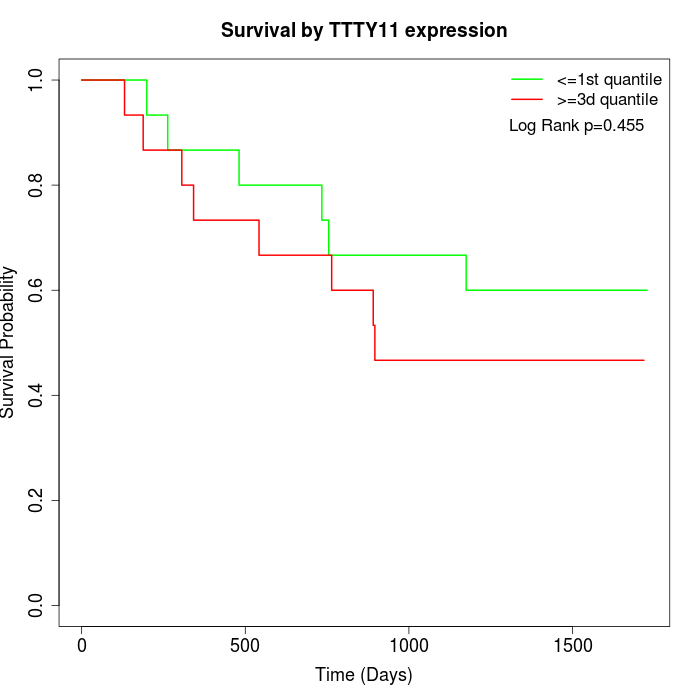
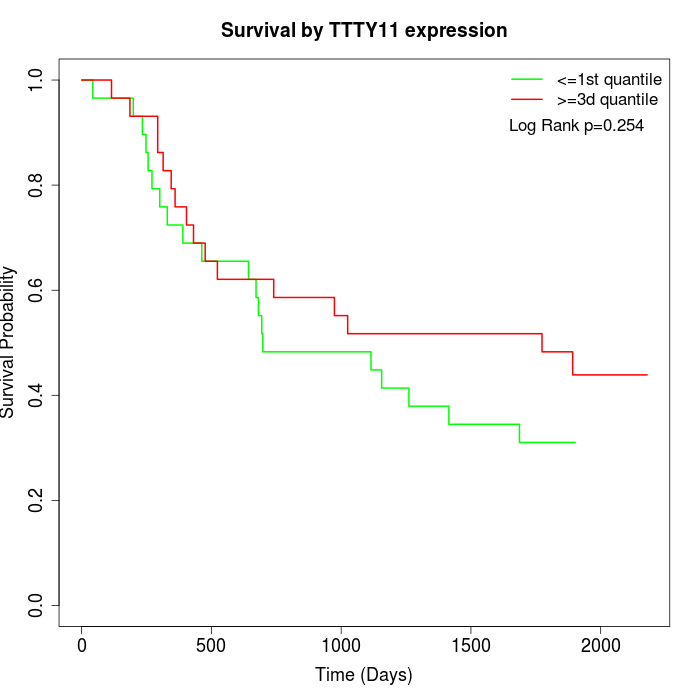
Survival by TTTY11 expression:
Note: Click image to view full size file.
Copy number change of TTTY11:
No record found for this gene.
Somatic mutations of TTTY11:
Generating mutation plots.
Highly correlated genes for TTTY11:
Showing top 20/303 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TTTY11 | FNDC9 | 0.748345 | 3 | 0 | 3 |
| TTTY11 | LINC01282 | 0.712599 | 3 | 0 | 3 |
| TTTY11 | LINC01210 | 0.710061 | 3 | 0 | 3 |
| TTTY11 | PRSS58 | 0.699379 | 4 | 0 | 4 |
| TTTY11 | ZNF169 | 0.689992 | 4 | 0 | 4 |
| TTTY11 | HOXB-AS3 | 0.683167 | 4 | 0 | 4 |
| TTTY11 | SPATA32 | 0.682081 | 5 | 0 | 4 |
| TTTY11 | MCHR2 | 0.678832 | 3 | 0 | 3 |
| TTTY11 | RIBC1 | 0.67781 | 3 | 0 | 3 |
| TTTY11 | CLYBL-AS2 | 0.67355 | 3 | 0 | 3 |
| TTTY11 | AGXT | 0.673374 | 3 | 0 | 3 |
| TTTY11 | SPERT | 0.673129 | 3 | 0 | 3 |
| TTTY11 | TESPA1 | 0.666813 | 3 | 0 | 3 |
| TTTY11 | PRAP1 | 0.6639 | 3 | 0 | 3 |
| TTTY11 | OR4D2 | 0.662778 | 6 | 0 | 5 |
| TTTY11 | CCDC154 | 0.659302 | 5 | 0 | 4 |
| TTTY11 | ALPI | 0.650932 | 3 | 0 | 3 |
| TTTY11 | CORIN | 0.649034 | 3 | 0 | 3 |
| TTTY11 | ACRV1 | 0.648845 | 3 | 0 | 3 |
| TTTY11 | MAPK4 | 0.64786 | 3 | 0 | 3 |
For details and further investigation, click here