| Full name: zinc finger and SCAN domain containing 1 | Alias Symbol: FLJ33779|ZNF915 | ||
| Type: protein-coding gene | Cytoband: 19q13.43 | ||
| Entrez ID: 284312 | HGNC ID: HGNC:23712 | Ensembl Gene: ENSG00000152467 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of ZSCAN1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ZSCAN1 | 284312 | 1562651_at | -0.1081 | 0.6858 | |
| GSE26886 | ZSCAN1 | 284312 | 1562651_at | -0.0917 | 0.3692 | |
| GSE45670 | ZSCAN1 | 284312 | 1562651_at | 0.1156 | 0.2814 | |
| GSE53622 | ZSCAN1 | 284312 | 118978 | 1.1089 | 0.0000 | |
| GSE53624 | ZSCAN1 | 284312 | 118978 | 0.7511 | 0.0000 | |
| GSE63941 | ZSCAN1 | 284312 | 1562651_at | 0.1638 | 0.3646 | |
| GSE77861 | ZSCAN1 | 284312 | 1562651_at | -0.1654 | 0.0687 | |
| GSE97050 | ZSCAN1 | 284312 | A_33_P3277826 | 0.2856 | 0.6114 | |
| TCGA | ZSCAN1 | 284312 | RNAseq | -1.1600 | 0.1193 |
Upregulated datasets: 1; Downregulated datasets: 0.
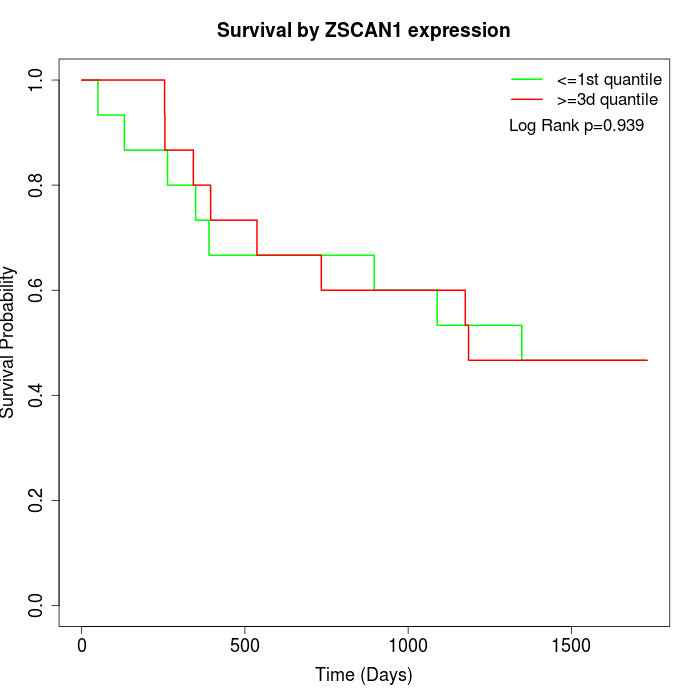
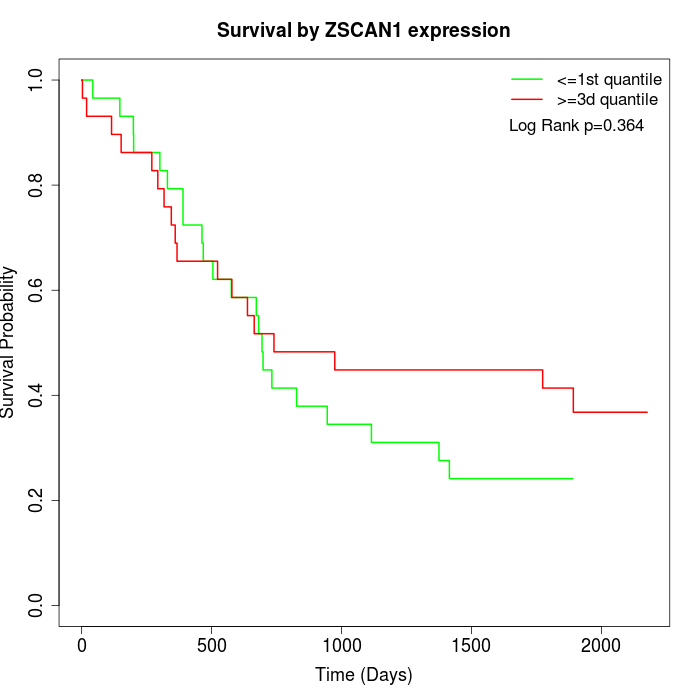
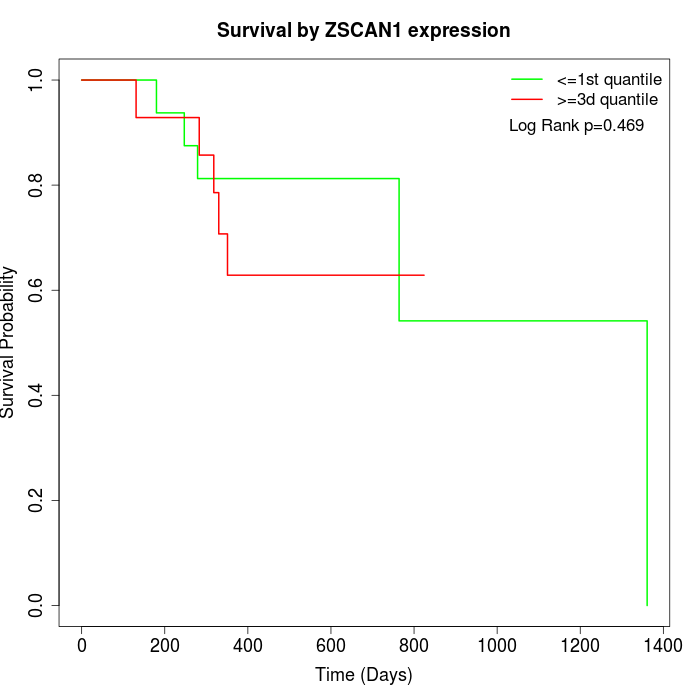
Survival by ZSCAN1 expression:
Note: Click image to view full size file.
Copy number change of ZSCAN1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ZSCAN1 | 284312 | 2 | 4 | 24 | |
| GSE20123 | ZSCAN1 | 284312 | 2 | 3 | 25 | |
| GSE43470 | ZSCAN1 | 284312 | 3 | 9 | 31 | |
| GSE46452 | ZSCAN1 | 284312 | 45 | 1 | 13 | |
| GSE47630 | ZSCAN1 | 284312 | 8 | 6 | 26 | |
| GSE54993 | ZSCAN1 | 284312 | 18 | 4 | 48 | |
| GSE54994 | ZSCAN1 | 284312 | 5 | 12 | 36 | |
| GSE60625 | ZSCAN1 | 284312 | 9 | 0 | 2 | |
| GSE74703 | ZSCAN1 | 284312 | 3 | 6 | 27 | |
| GSE74704 | ZSCAN1 | 284312 | 2 | 1 | 17 | |
| TCGA | ZSCAN1 | 284312 | 20 | 15 | 61 |
Total number of gains: 117; Total number of losses: 61; Total Number of normals: 310.
Somatic mutations of ZSCAN1:
Generating mutation plots.
Highly correlated genes for ZSCAN1:
Showing top 20/125 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ZSCAN1 | VGLL2 | 0.691102 | 3 | 0 | 3 |
| ZSCAN1 | LINC01012 | 0.678102 | 3 | 0 | 3 |
| ZSCAN1 | SIGLEC6 | 0.669615 | 3 | 0 | 3 |
| ZSCAN1 | TRIM3 | 0.669107 | 3 | 0 | 3 |
| ZSCAN1 | LINC00315 | 0.662177 | 4 | 0 | 4 |
| ZSCAN1 | NRSN2 | 0.658052 | 3 | 0 | 3 |
| ZSCAN1 | KIF12 | 0.648248 | 4 | 0 | 4 |
| ZSCAN1 | GPR37L1 | 0.638026 | 4 | 0 | 3 |
| ZSCAN1 | ANKUB1 | 0.629594 | 3 | 0 | 3 |
| ZSCAN1 | CRISP1 | 0.622062 | 4 | 0 | 4 |
| ZSCAN1 | CLYBL-AS1 | 0.620652 | 3 | 0 | 3 |
| ZSCAN1 | TAS2R43 | 0.618091 | 3 | 0 | 3 |
| ZSCAN1 | SPDYE3 | 0.616708 | 3 | 0 | 3 |
| ZSCAN1 | BTN1A1 | 0.611925 | 4 | 0 | 4 |
| ZSCAN1 | STARD7-AS1 | 0.611882 | 3 | 0 | 3 |
| ZSCAN1 | APOA5 | 0.610189 | 5 | 0 | 4 |
| ZSCAN1 | SYN3 | 0.609901 | 5 | 0 | 4 |
| ZSCAN1 | LRRTM2 | 0.609864 | 4 | 0 | 3 |
| ZSCAN1 | LINC01210 | 0.603694 | 4 | 0 | 3 |
| ZSCAN1 | HPYR1 | 0.59861 | 3 | 0 | 3 |
For details and further investigation, click here