| Full name: adaptor related protein complex 1 subunit sigma 2 | Alias Symbol: SIGMA1B | ||
| Type: protein-coding gene | Cytoband: Xp22.2 | ||
| Entrez ID: 8905 | HGNC ID: HGNC:560 | Ensembl Gene: ENSG00000182287 | OMIM ID: 300629 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of AP1S2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | AP1S2 | 8905 | 230264_s_at | -0.8472 | 0.4050 | |
| GSE20347 | AP1S2 | 8905 | 203300_x_at | 0.4881 | 0.0755 | |
| GSE23400 | AP1S2 | 8905 | 203300_x_at | -0.0570 | 0.7285 | |
| GSE26886 | AP1S2 | 8905 | 228415_at | -0.0573 | 0.8724 | |
| GSE29001 | AP1S2 | 8905 | 203300_x_at | 0.2067 | 0.5925 | |
| GSE38129 | AP1S2 | 8905 | 203300_x_at | 0.0026 | 0.9938 | |
| GSE45670 | AP1S2 | 8905 | 228415_at | -0.1813 | 0.4696 | |
| GSE53622 | AP1S2 | 8905 | 59018 | -0.4919 | 0.0000 | |
| GSE53624 | AP1S2 | 8905 | 45243 | -0.0173 | 0.9183 | |
| GSE63941 | AP1S2 | 8905 | 230264_s_at | -1.8993 | 0.0377 | |
| GSE77861 | AP1S2 | 8905 | 228415_at | 0.3073 | 0.7216 | |
| GSE97050 | AP1S2 | 8905 | A_33_P3258782 | -0.7267 | 0.1596 | |
| SRP007169 | AP1S2 | 8905 | RNAseq | 0.7763 | 0.0645 | |
| SRP008496 | AP1S2 | 8905 | RNAseq | 1.2768 | 0.0008 | |
| SRP064894 | AP1S2 | 8905 | RNAseq | 0.2925 | 0.2726 | |
| SRP133303 | AP1S2 | 8905 | RNAseq | 0.3837 | 0.2186 | |
| SRP159526 | AP1S2 | 8905 | RNAseq | 0.3036 | 0.5217 | |
| SRP193095 | AP1S2 | 8905 | RNAseq | 0.1346 | 0.2977 | |
| SRP219564 | AP1S2 | 8905 | RNAseq | 0.0040 | 0.9962 | |
| TCGA | AP1S2 | 8905 | RNAseq | -0.1152 | 0.2586 |
Upregulated datasets: 1; Downregulated datasets: 1.
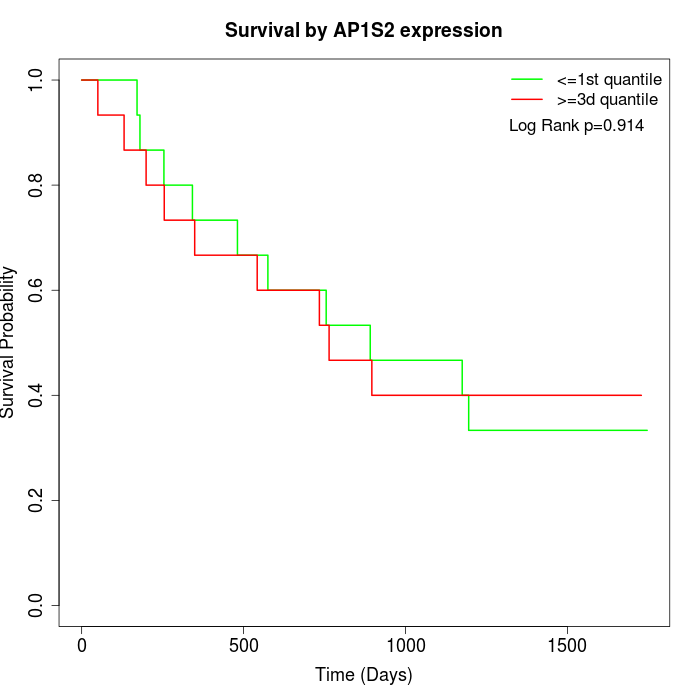
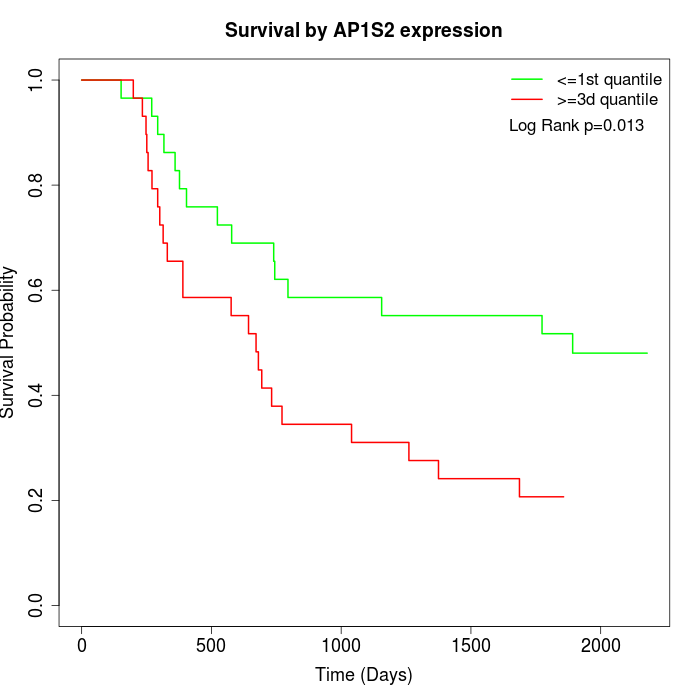
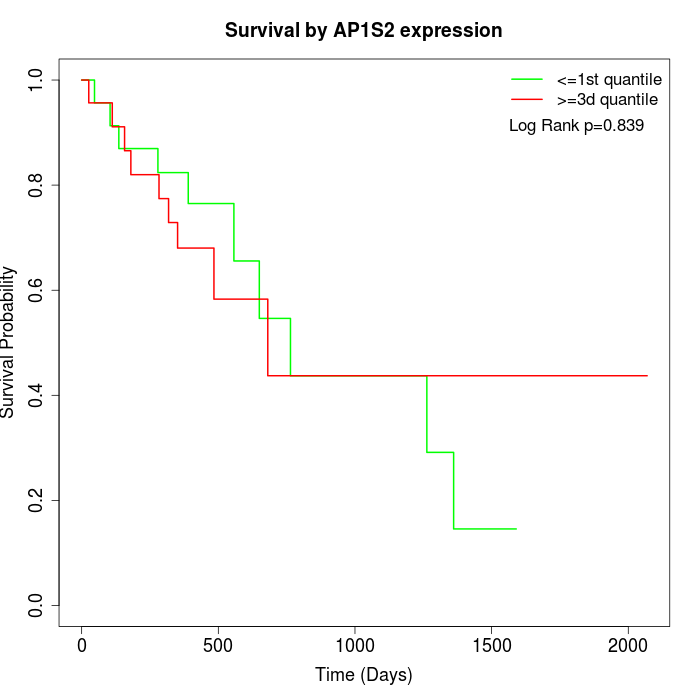
Survival by AP1S2 expression:
Note: Click image to view full size file.
Copy number change of AP1S2:
No record found for this gene.
Somatic mutations of AP1S2:
Generating mutation plots.
Highly correlated genes for AP1S2:
Showing top 20/872 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| AP1S2 | MYADM | 0.80508 | 3 | 0 | 3 |
| AP1S2 | FIBIN | 0.799963 | 3 | 0 | 3 |
| AP1S2 | MAP1B | 0.792018 | 6 | 0 | 6 |
| AP1S2 | RHOJ | 0.791834 | 4 | 0 | 4 |
| AP1S2 | BBS1 | 0.780299 | 5 | 0 | 5 |
| AP1S2 | MYOCD | 0.777884 | 4 | 0 | 4 |
| AP1S2 | ZEB1 | 0.768759 | 3 | 0 | 3 |
| AP1S2 | ILK | 0.768642 | 6 | 0 | 6 |
| AP1S2 | MSRB3 | 0.766912 | 6 | 0 | 6 |
| AP1S2 | PYGM | 0.763191 | 5 | 0 | 5 |
| AP1S2 | GNPDA2 | 0.76229 | 5 | 0 | 5 |
| AP1S2 | TMOD2 | 0.761643 | 4 | 0 | 4 |
| AP1S2 | SYNC | 0.760104 | 6 | 0 | 6 |
| AP1S2 | BVES | 0.759215 | 5 | 0 | 4 |
| AP1S2 | ANO6 | 0.759116 | 4 | 0 | 4 |
| AP1S2 | TCEAL6 | 0.752233 | 3 | 0 | 3 |
| AP1S2 | FOXN3 | 0.751356 | 4 | 0 | 3 |
| AP1S2 | LRRN4CL | 0.750141 | 5 | 0 | 5 |
| AP1S2 | C20orf194 | 0.747116 | 3 | 0 | 3 |
| AP1S2 | HMCN1 | 0.744963 | 7 | 0 | 7 |
For details and further investigation, click here