| Full name: glycogen phosphorylase, muscle associated | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 11q13.1 | ||
| Entrez ID: 5837 | HGNC ID: HGNC:9726 | Ensembl Gene: ENSG00000068976 | OMIM ID: 608455 |
| Drug and gene relationship at DGIdb | |||
PYGM involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04910 | Insulin signaling pathway | |
| hsa04922 | Glucagon signaling pathway | |
| hsa04931 | Insulin resistance |
Expression of PYGM:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PYGM | 5837 | 205577_at | -1.0809 | 0.3662 | |
| GSE20347 | PYGM | 5837 | 205577_at | -0.2215 | 0.1051 | |
| GSE23400 | PYGM | 5837 | 205577_at | -0.6469 | 0.0000 | |
| GSE26886 | PYGM | 5837 | 205577_at | 0.1099 | 0.4557 | |
| GSE29001 | PYGM | 5837 | 205577_at | -0.2730 | 0.0226 | |
| GSE38129 | PYGM | 5837 | 205577_at | -0.9134 | 0.0005 | |
| GSE45670 | PYGM | 5837 | 205577_at | -0.6576 | 0.0001 | |
| GSE53622 | PYGM | 5837 | 60647 | -2.2462 | 0.0000 | |
| GSE53624 | PYGM | 5837 | 60647 | -1.9938 | 0.0000 | |
| GSE63941 | PYGM | 5837 | 205577_at | 0.1512 | 0.4261 | |
| GSE77861 | PYGM | 5837 | 205577_at | -0.1178 | 0.2928 | |
| GSE97050 | PYGM | 5837 | A_23_P405815 | -1.9031 | 0.1057 | |
| SRP064894 | PYGM | 5837 | RNAseq | -1.2007 | 0.0045 | |
| SRP133303 | PYGM | 5837 | RNAseq | -3.0421 | 0.0000 | |
| SRP159526 | PYGM | 5837 | RNAseq | -1.7019 | 0.0158 | |
| SRP219564 | PYGM | 5837 | RNAseq | -1.4562 | 0.2894 | |
| TCGA | PYGM | 5837 | RNAseq | -1.5541 | 0.0008 |
Upregulated datasets: 0; Downregulated datasets: 6.
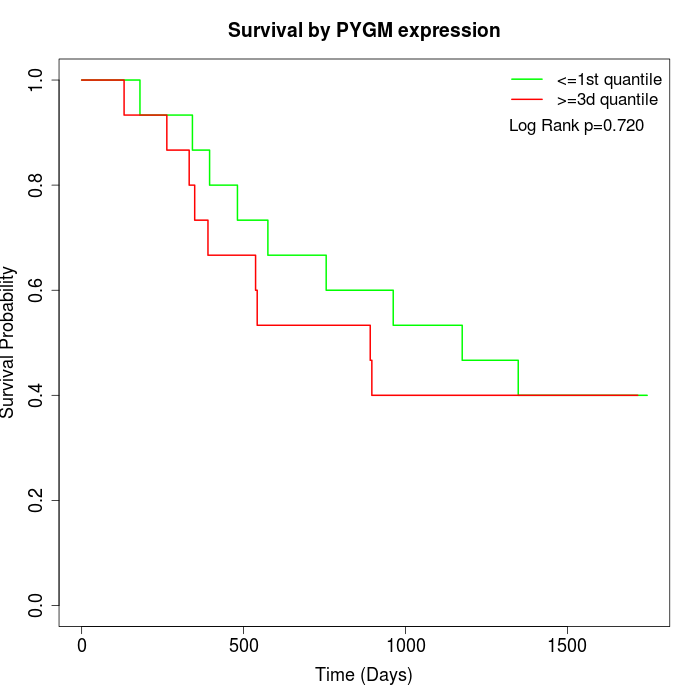
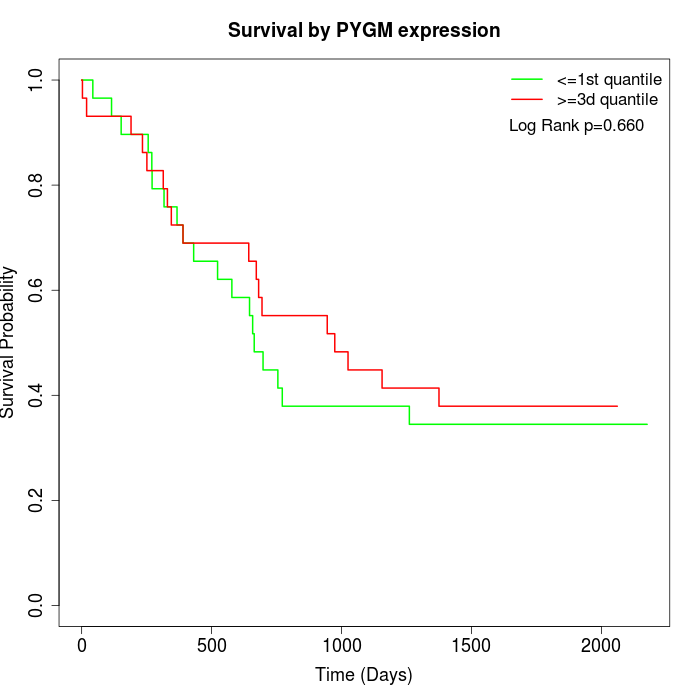
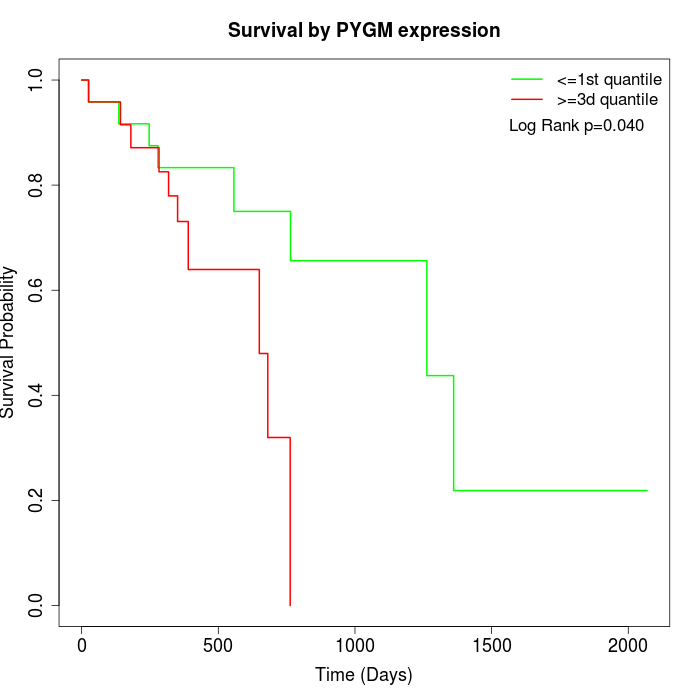
Survival by PYGM expression:
Note: Click image to view full size file.
Copy number change of PYGM:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PYGM | 5837 | 7 | 6 | 17 | |
| GSE20123 | PYGM | 5837 | 7 | 6 | 17 | |
| GSE43470 | PYGM | 5837 | 1 | 3 | 39 | |
| GSE46452 | PYGM | 5837 | 9 | 4 | 46 | |
| GSE47630 | PYGM | 5837 | 5 | 6 | 29 | |
| GSE54993 | PYGM | 5837 | 3 | 1 | 66 | |
| GSE54994 | PYGM | 5837 | 6 | 5 | 42 | |
| GSE60625 | PYGM | 5837 | 0 | 3 | 8 | |
| GSE74703 | PYGM | 5837 | 1 | 1 | 34 | |
| GSE74704 | PYGM | 5837 | 5 | 4 | 11 | |
| TCGA | PYGM | 5837 | 20 | 8 | 68 |
Total number of gains: 64; Total number of losses: 47; Total Number of normals: 377.
Somatic mutations of PYGM:
Generating mutation plots.
Highly correlated genes for PYGM:
Showing top 20/1278 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PYGM | UBXN10 | 0.880213 | 4 | 0 | 4 |
| PYGM | CYP21A2 | 0.873736 | 3 | 0 | 3 |
| PYGM | CLEC3B | 0.846305 | 3 | 0 | 3 |
| PYGM | ADCY5 | 0.841694 | 5 | 0 | 5 |
| PYGM | LPP | 0.832145 | 7 | 0 | 7 |
| PYGM | SLMAP | 0.831995 | 5 | 0 | 5 |
| PYGM | CASQ2 | 0.82983 | 10 | 0 | 10 |
| PYGM | HSPB7 | 0.826642 | 9 | 0 | 9 |
| PYGM | PPP1R12C | 0.821222 | 3 | 0 | 3 |
| PYGM | HPSE2 | 0.817985 | 9 | 0 | 9 |
| PYGM | PRR26 | 0.815544 | 4 | 0 | 4 |
| PYGM | SH3BGR | 0.813455 | 8 | 0 | 8 |
| PYGM | SYNM | 0.811364 | 9 | 0 | 9 |
| PYGM | NACC2 | 0.809494 | 7 | 0 | 7 |
| PYGM | RBFOX3 | 0.807659 | 3 | 0 | 3 |
| PYGM | PDZRN4 | 0.807483 | 9 | 0 | 8 |
| PYGM | XKR4 | 0.802542 | 6 | 0 | 6 |
| PYGM | NEGR1 | 0.799388 | 6 | 0 | 6 |
| PYGM | ACTBL2 | 0.797884 | 3 | 0 | 3 |
| PYGM | VMAC | 0.797442 | 3 | 0 | 3 |
For details and further investigation, click here