| Full name: zinc finger E-box binding homeobox 1 | Alias Symbol: BZP|ZEB|AREB6|NIL-2-A|Zfhep|Zfhx1a|FECD6 | ||
| Type: protein-coding gene | Cytoband: 10p11.22 | ||
| Entrez ID: 6935 | HGNC ID: HGNC:11642 | Ensembl Gene: ENSG00000148516 | OMIM ID: 189909 |
| Drug and gene relationship at DGIdb | |||
Expression of ZEB1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | ZEB1 | 6935 | 88592 | -0.2934 | 0.0178 | |
| GSE53624 | ZEB1 | 6935 | 88592 | -0.1949 | 0.0365 | |
| GSE97050 | ZEB1 | 6935 | A_33_P3640690 | -0.8229 | 0.1298 | |
| SRP007169 | ZEB1 | 6935 | RNAseq | 2.3167 | 0.0000 | |
| SRP008496 | ZEB1 | 6935 | RNAseq | 2.8469 | 0.0000 | |
| SRP064894 | ZEB1 | 6935 | RNAseq | 0.5084 | 0.1555 | |
| SRP133303 | ZEB1 | 6935 | RNAseq | 0.2610 | 0.4324 | |
| SRP159526 | ZEB1 | 6935 | RNAseq | -0.0438 | 0.9020 | |
| SRP193095 | ZEB1 | 6935 | RNAseq | 1.2129 | 0.0000 | |
| SRP219564 | ZEB1 | 6935 | RNAseq | 0.0707 | 0.9217 | |
| TCGA | ZEB1 | 6935 | RNAseq | -0.3342 | 0.0033 |
Upregulated datasets: 3; Downregulated datasets: 0.
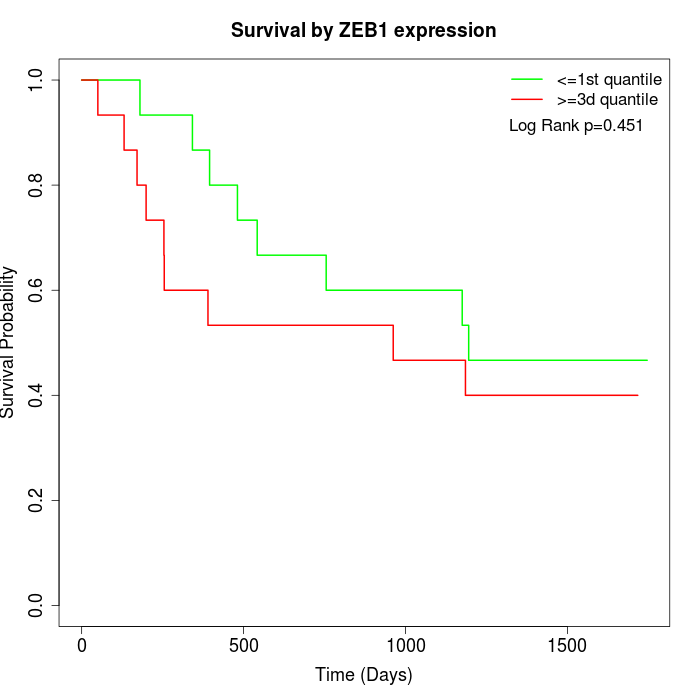
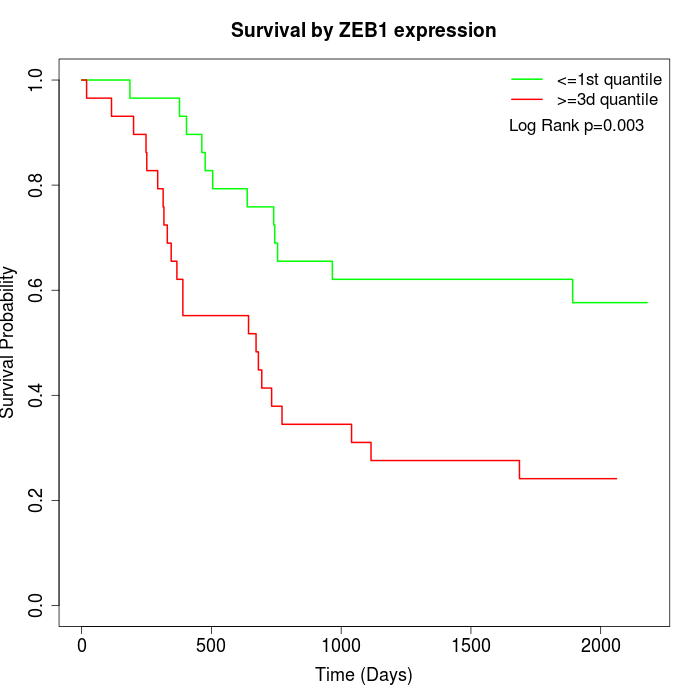
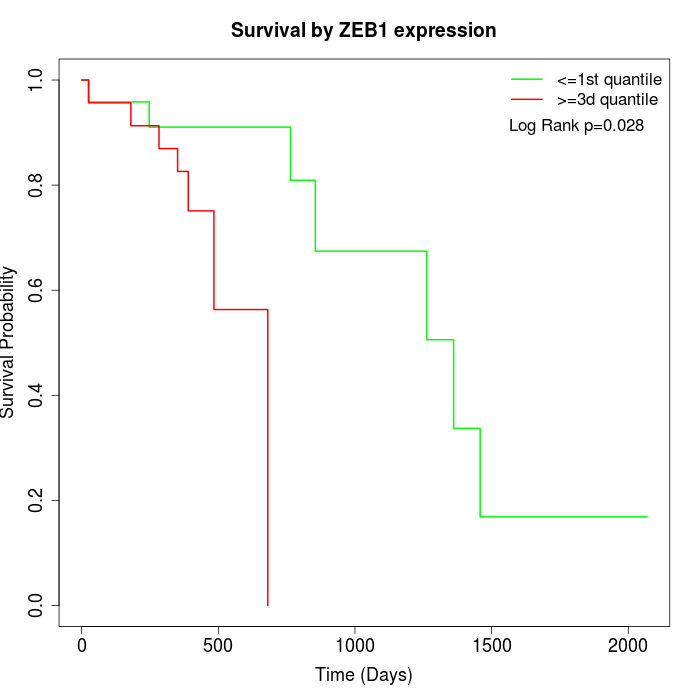
Survival by ZEB1 expression:
Note: Click image to view full size file.
Copy number change of ZEB1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ZEB1 | 6935 | 5 | 7 | 18 | |
| GSE20123 | ZEB1 | 6935 | 5 | 6 | 19 | |
| GSE43470 | ZEB1 | 6935 | 4 | 3 | 36 | |
| GSE46452 | ZEB1 | 6935 | 1 | 14 | 44 | |
| GSE47630 | ZEB1 | 6935 | 5 | 15 | 20 | |
| GSE54993 | ZEB1 | 6935 | 9 | 0 | 61 | |
| GSE54994 | ZEB1 | 6935 | 3 | 9 | 41 | |
| GSE60625 | ZEB1 | 6935 | 0 | 0 | 11 | |
| GSE74703 | ZEB1 | 6935 | 2 | 2 | 32 | |
| GSE74704 | ZEB1 | 6935 | 0 | 5 | 15 | |
| TCGA | ZEB1 | 6935 | 18 | 24 | 54 |
Total number of gains: 52; Total number of losses: 85; Total Number of normals: 351.
Somatic mutations of ZEB1:
Generating mutation plots.
Highly correlated genes for ZEB1:
Showing top 20/255 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ZEB1 | TGFB1I1 | 0.862808 | 3 | 0 | 3 |
| ZEB1 | KANK2 | 0.853868 | 3 | 0 | 3 |
| ZEB1 | PDLIM3 | 0.851997 | 3 | 0 | 3 |
| ZEB1 | MAP1A | 0.841796 | 3 | 0 | 3 |
| ZEB1 | FERMT2 | 0.838034 | 3 | 0 | 3 |
| ZEB1 | ZCCHC24 | 0.828782 | 3 | 0 | 3 |
| ZEB1 | FILIP1L | 0.823128 | 3 | 0 | 3 |
| ZEB1 | ILK | 0.81873 | 3 | 0 | 3 |
| ZEB1 | CLIP3 | 0.818344 | 3 | 0 | 3 |
| ZEB1 | RUNX1T1 | 0.817802 | 3 | 0 | 3 |
| ZEB1 | PARVA | 0.816316 | 3 | 0 | 3 |
| ZEB1 | PKIG | 0.815987 | 3 | 0 | 3 |
| ZEB1 | DDR2 | 0.815514 | 3 | 0 | 3 |
| ZEB1 | CALD1 | 0.809301 | 3 | 0 | 3 |
| ZEB1 | ITGA1 | 0.80879 | 3 | 0 | 3 |
| ZEB1 | JAM3 | 0.808387 | 3 | 0 | 3 |
| ZEB1 | SGCB | 0.80621 | 3 | 0 | 3 |
| ZEB1 | ANTXR2 | 0.801915 | 3 | 0 | 3 |
| ZEB1 | SGCD | 0.8007 | 3 | 0 | 3 |
| ZEB1 | MRGPRF | 0.800101 | 3 | 0 | 3 |
For details and further investigation, click here