| Full name: arrestin beta 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 11q13.4 | ||
| Entrez ID: 408 | HGNC ID: HGNC:711 | Ensembl Gene: ENSG00000137486 | OMIM ID: 107940 |
| Drug and gene relationship at DGIdb | |||
ARRB1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04010 | MAPK signaling pathway | |
| hsa04062 | Chemokine signaling pathway | |
| hsa04340 | Hedgehog signaling pathway | |
| hsa04740 | Olfactory transduction | |
| hsa05032 | Morphine addiction |
Expression of ARRB1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ARRB1 | 408 | 221861_at | -0.4184 | 0.3268 | |
| GSE20347 | ARRB1 | 408 | 221861_at | -0.2233 | 0.0061 | |
| GSE23400 | ARRB1 | 408 | 221861_at | -0.3759 | 0.0000 | |
| GSE26886 | ARRB1 | 408 | 43511_s_at | -0.8397 | 0.0123 | |
| GSE29001 | ARRB1 | 408 | 221861_at | -0.3556 | 0.0333 | |
| GSE38129 | ARRB1 | 408 | 221861_at | -0.6546 | 0.0000 | |
| GSE45670 | ARRB1 | 408 | 221861_at | -0.3793 | 0.0038 | |
| GSE53622 | ARRB1 | 408 | 26372 | -1.1817 | 0.0000 | |
| GSE53624 | ARRB1 | 408 | 26372 | -0.8902 | 0.0000 | |
| GSE63941 | ARRB1 | 408 | 221861_at | -0.8403 | 0.1654 | |
| GSE77861 | ARRB1 | 408 | 221861_at | -0.3264 | 0.0077 | |
| GSE97050 | ARRB1 | 408 | A_24_P386622 | -0.9260 | 0.1506 | |
| SRP007169 | ARRB1 | 408 | RNAseq | -0.6055 | 0.3397 | |
| SRP008496 | ARRB1 | 408 | RNAseq | -0.9488 | 0.0107 | |
| SRP064894 | ARRB1 | 408 | RNAseq | -0.3837 | 0.2349 | |
| SRP133303 | ARRB1 | 408 | RNAseq | -1.1801 | 0.0001 | |
| SRP159526 | ARRB1 | 408 | RNAseq | -1.4782 | 0.0000 | |
| SRP193095 | ARRB1 | 408 | RNAseq | -0.6318 | 0.0087 | |
| SRP219564 | ARRB1 | 408 | RNAseq | -0.7912 | 0.3246 | |
| TCGA | ARRB1 | 408 | RNAseq | -1.0074 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 4.
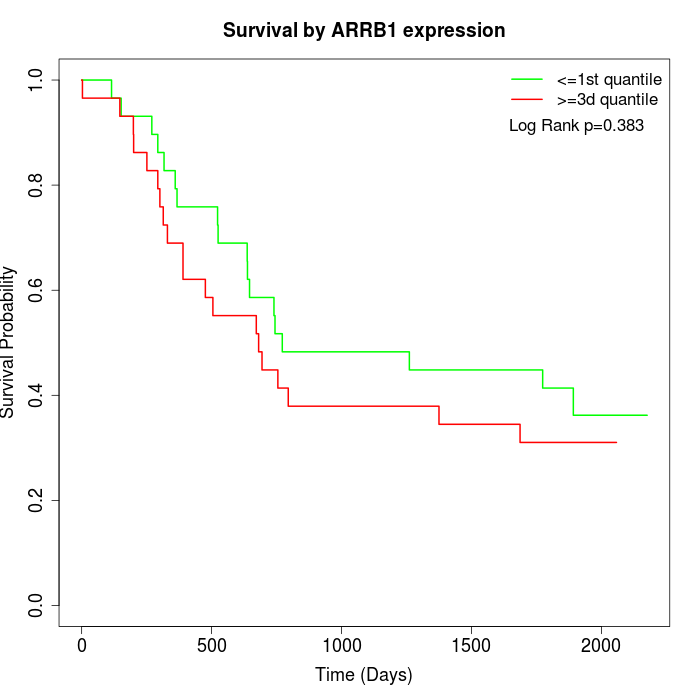
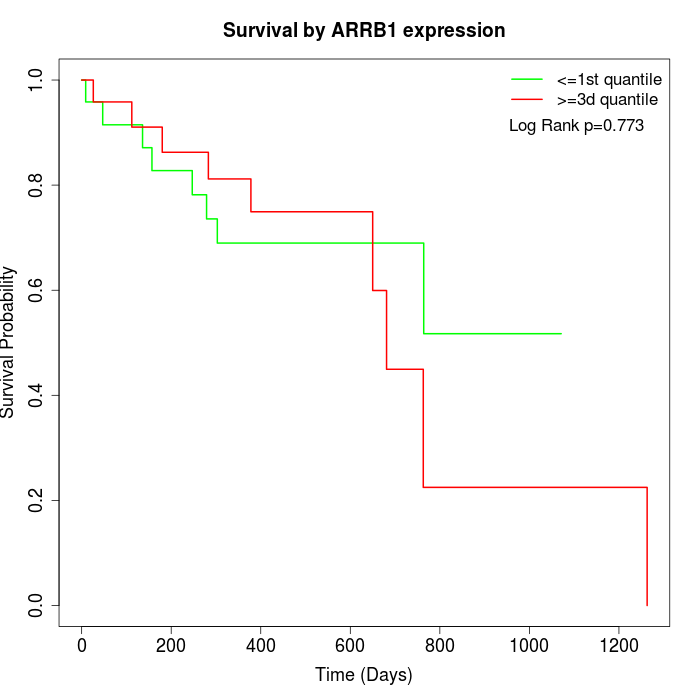
Survival by ARRB1 expression:
Note: Click image to view full size file.
Copy number change of ARRB1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ARRB1 | 408 | 6 | 8 | 16 | |
| GSE20123 | ARRB1 | 408 | 6 | 7 | 17 | |
| GSE43470 | ARRB1 | 408 | 5 | 4 | 34 | |
| GSE46452 | ARRB1 | 408 | 7 | 22 | 30 | |
| GSE47630 | ARRB1 | 408 | 6 | 14 | 20 | |
| GSE54993 | ARRB1 | 408 | 8 | 2 | 60 | |
| GSE54994 | ARRB1 | 408 | 7 | 16 | 30 | |
| GSE60625 | ARRB1 | 408 | 4 | 3 | 4 | |
| GSE74703 | ARRB1 | 408 | 4 | 3 | 29 | |
| GSE74704 | ARRB1 | 408 | 4 | 3 | 13 | |
| TCGA | ARRB1 | 408 | 21 | 32 | 43 |
Total number of gains: 78; Total number of losses: 114; Total Number of normals: 296.
Somatic mutations of ARRB1:
Generating mutation plots.
Highly correlated genes for ARRB1:
Showing top 20/1272 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ARRB1 | TNFSF12 | 0.826517 | 3 | 0 | 3 |
| ARRB1 | SORCS1 | 0.818834 | 4 | 0 | 4 |
| ARRB1 | RASSF3 | 0.811957 | 4 | 0 | 4 |
| ARRB1 | GRHPR | 0.803262 | 3 | 0 | 3 |
| ARRB1 | LRFN5 | 0.771347 | 5 | 0 | 5 |
| ARRB1 | ANO5 | 0.767518 | 5 | 0 | 5 |
| ARRB1 | HSPB6 | 0.759649 | 8 | 0 | 8 |
| ARRB1 | DNAJC15 | 0.756475 | 3 | 0 | 3 |
| ARRB1 | RNASE4 | 0.754733 | 5 | 0 | 5 |
| ARRB1 | ZBTB22 | 0.753186 | 3 | 0 | 3 |
| ARRB1 | GPRASP2 | 0.748124 | 3 | 0 | 3 |
| ARRB1 | KIAA1328 | 0.746583 | 4 | 0 | 4 |
| ARRB1 | AFMID | 0.740152 | 3 | 0 | 3 |
| ARRB1 | MBNL1-AS1 | 0.73483 | 3 | 0 | 3 |
| ARRB1 | STARD9 | 0.733329 | 4 | 0 | 4 |
| ARRB1 | SGCA | 0.733086 | 9 | 0 | 9 |
| ARRB1 | C5orf24 | 0.731453 | 4 | 0 | 4 |
| ARRB1 | NEK9 | 0.730924 | 3 | 0 | 3 |
| ARRB1 | HOXA4 | 0.73055 | 7 | 0 | 6 |
| ARRB1 | C1QTNF7 | 0.729097 | 6 | 0 | 5 |
For details and further investigation, click here