| Full name: leucine rich repeat and fibronectin type III domain containing 5 | Alias Symbol: FIGLER8|SALM5 | ||
| Type: protein-coding gene | Cytoband: 14q21.1 | ||
| Entrez ID: 145581 | HGNC ID: HGNC:20360 | Ensembl Gene: ENSG00000165379 | OMIM ID: 612811 |
| Drug and gene relationship at DGIdb | |||
Expression of LRFN5:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LRFN5 | 145581 | 230644_at | -0.4904 | 0.1302 | |
| GSE26886 | LRFN5 | 145581 | 230644_at | 0.7255 | 0.0589 | |
| GSE53622 | LRFN5 | 145581 | 18592 | -1.6936 | 0.0000 | |
| GSE53624 | LRFN5 | 145581 | 18592 | -1.3572 | 0.0000 | |
| GSE63941 | LRFN5 | 145581 | 230644_at | -0.7477 | 0.0305 | |
| GSE97050 | LRFN5 | 145581 | A_23_P163195 | -0.4214 | 0.2144 | |
| SRP133303 | LRFN5 | 145581 | RNAseq | -1.3961 | 0.0000 | |
| SRP159526 | LRFN5 | 145581 | RNAseq | -1.0250 | 0.0752 | |
| SRP219564 | LRFN5 | 145581 | RNAseq | -0.3613 | 0.7010 | |
| TCGA | LRFN5 | 145581 | RNAseq | -0.9851 | 0.0236 |
Upregulated datasets: 0; Downregulated datasets: 3.
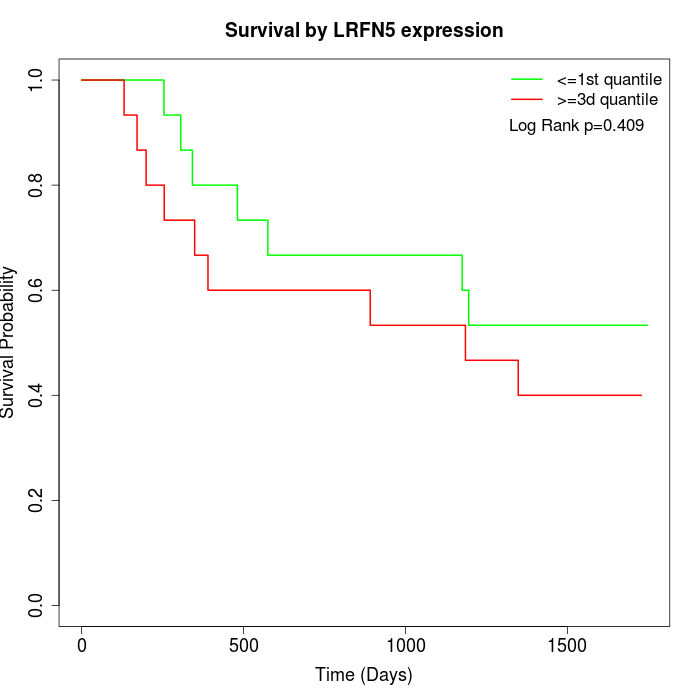
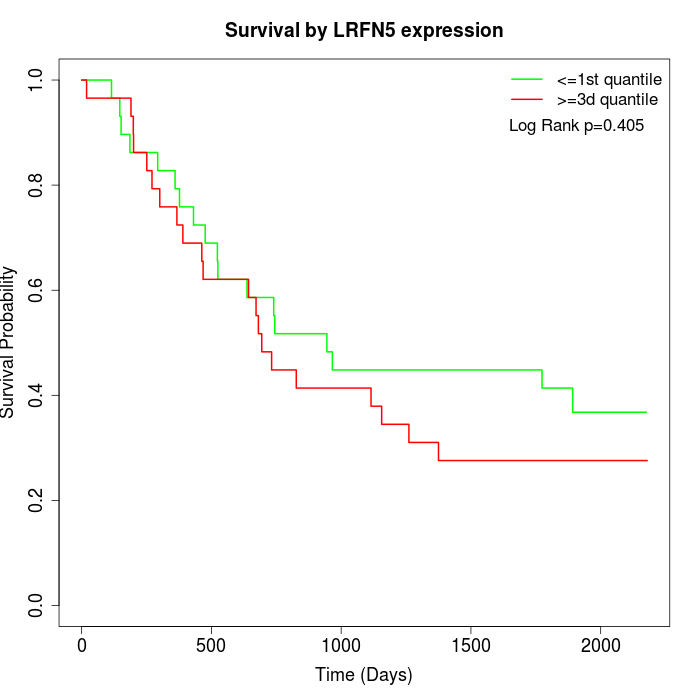
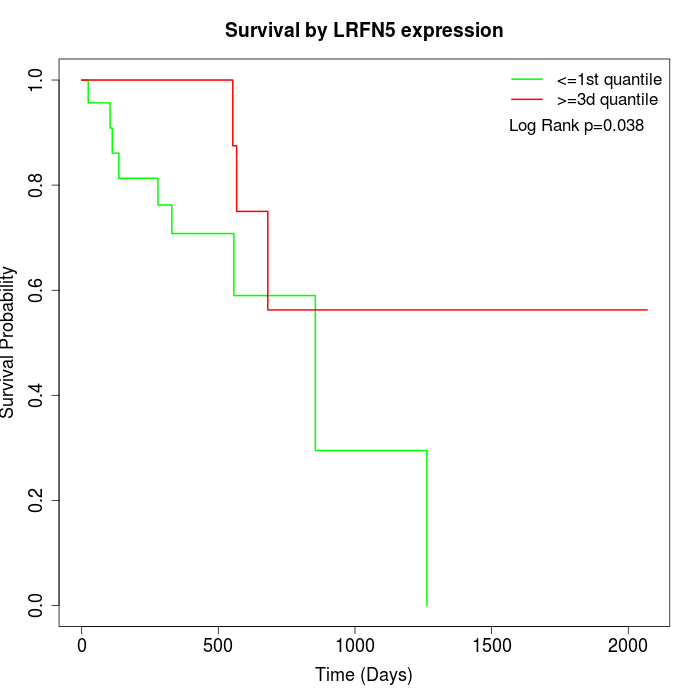
Survival by LRFN5 expression:
Note: Click image to view full size file.
Copy number change of LRFN5:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LRFN5 | 145581 | 7 | 5 | 18 | |
| GSE20123 | LRFN5 | 145581 | 7 | 5 | 18 | |
| GSE43470 | LRFN5 | 145581 | 8 | 3 | 32 | |
| GSE46452 | LRFN5 | 145581 | 18 | 2 | 39 | |
| GSE47630 | LRFN5 | 145581 | 11 | 10 | 19 | |
| GSE54993 | LRFN5 | 145581 | 3 | 9 | 58 | |
| GSE54994 | LRFN5 | 145581 | 18 | 5 | 30 | |
| GSE60625 | LRFN5 | 145581 | 0 | 2 | 9 | |
| GSE74703 | LRFN5 | 145581 | 7 | 3 | 26 | |
| GSE74704 | LRFN5 | 145581 | 4 | 3 | 13 | |
| TCGA | LRFN5 | 145581 | 31 | 14 | 51 |
Total number of gains: 114; Total number of losses: 61; Total Number of normals: 313.
Somatic mutations of LRFN5:
Generating mutation plots.
Highly correlated genes for LRFN5:
Showing top 20/773 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LRFN5 | CASQ2 | 0.843755 | 4 | 0 | 4 |
| LRFN5 | OGN | 0.839031 | 5 | 0 | 5 |
| LRFN5 | MSRB3 | 0.834741 | 4 | 0 | 4 |
| LRFN5 | PDE1A | 0.825343 | 5 | 0 | 5 |
| LRFN5 | SORCS1 | 0.823723 | 4 | 0 | 4 |
| LRFN5 | ITPR1 | 0.822271 | 4 | 0 | 4 |
| LRFN5 | TRAK1 | 0.820635 | 3 | 0 | 3 |
| LRFN5 | ASPA | 0.817568 | 5 | 0 | 5 |
| LRFN5 | FHL5 | 0.810172 | 4 | 0 | 4 |
| LRFN5 | AFF3 | 0.807125 | 3 | 0 | 3 |
| LRFN5 | SLIT3 | 0.803259 | 3 | 0 | 3 |
| LRFN5 | SLITRK3 | 0.802031 | 4 | 0 | 4 |
| LRFN5 | HPSE2 | 0.800107 | 4 | 0 | 4 |
| LRFN5 | TNFSF12 | 0.795083 | 3 | 0 | 3 |
| LRFN5 | ITGA9 | 0.794976 | 4 | 0 | 4 |
| LRFN5 | LDB3 | 0.794541 | 4 | 0 | 4 |
| LRFN5 | HSPB6 | 0.794503 | 4 | 0 | 4 |
| LRFN5 | TCEAL1 | 0.792381 | 4 | 0 | 4 |
| LRFN5 | APOLD1 | 0.790307 | 4 | 0 | 4 |
| LRFN5 | CFL2 | 0.789547 | 4 | 0 | 4 |
For details and further investigation, click here