| Full name: ankyrin repeat and SOCS box containing 3 | Alias Symbol: ASB-3 | ||
| Type: protein-coding gene | Cytoband: 2p16.2 | ||
| Entrez ID: 51130 | HGNC ID: HGNC:16013 | Ensembl Gene: ENSG00000115239 | OMIM ID: 605760 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of ASB3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | ASB3 | 51130 | 135683 | 0.6190 | 0.0000 | |
| GSE53624 | ASB3 | 51130 | 135683 | 0.6599 | 0.0000 | |
| GSE97050 | ASB3 | 51130 | A_23_P56833 | 0.1405 | 0.5890 | |
| SRP007169 | ASB3 | 51130 | RNAseq | -0.1868 | 0.8197 | |
| SRP008496 | ASB3 | 51130 | RNAseq | 0.3495 | 0.6552 | |
| SRP064894 | ASB3 | 51130 | RNAseq | 0.3754 | 0.0191 | |
| SRP133303 | ASB3 | 51130 | RNAseq | 0.1471 | 0.3477 | |
| SRP159526 | ASB3 | 51130 | RNAseq | 0.7509 | 0.0334 | |
| SRP193095 | ASB3 | 51130 | RNAseq | 0.1008 | 0.3731 | |
| SRP219564 | ASB3 | 51130 | RNAseq | -0.0560 | 0.8442 | |
| TCGA | ASB3 | 51130 | RNAseq | 0.2036 | 0.0010 |
Upregulated datasets: 0; Downregulated datasets: 0.
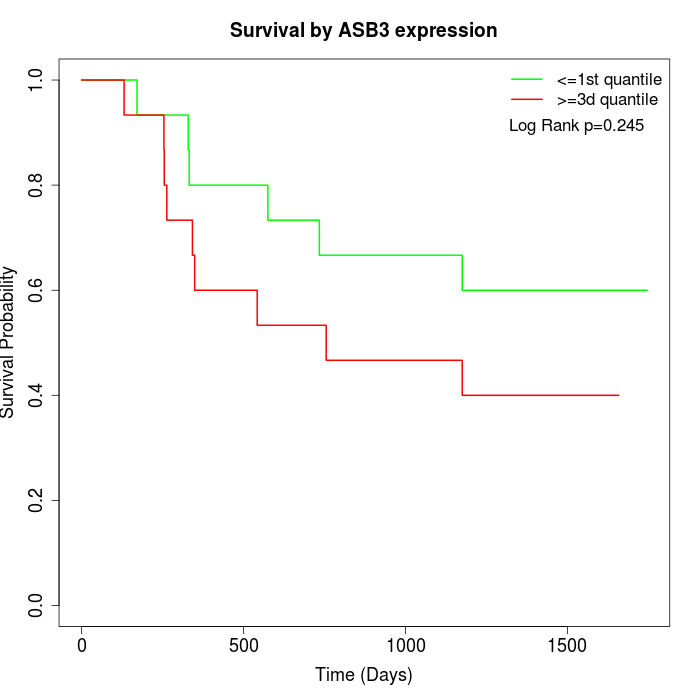
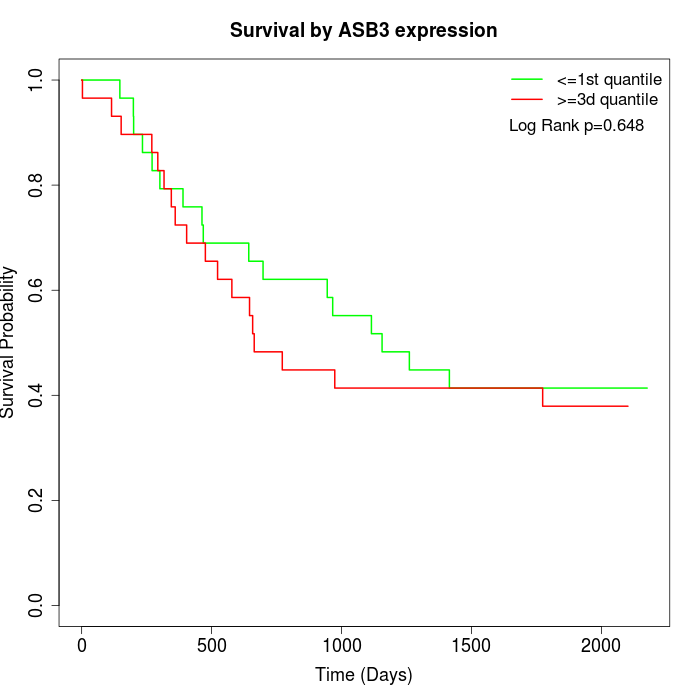
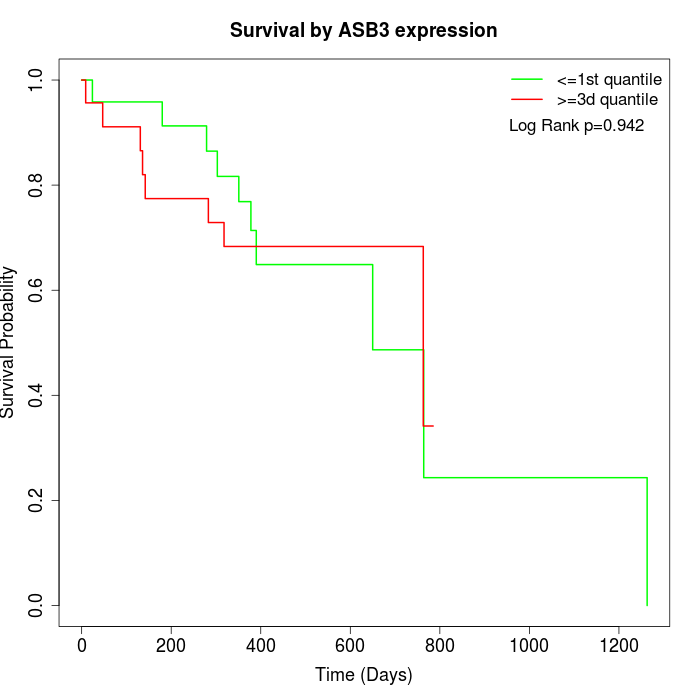
Survival by ASB3 expression:
Note: Click image to view full size file.
Copy number change of ASB3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ASB3 | 51130 | 8 | 0 | 22 | |
| GSE20123 | ASB3 | 51130 | 7 | 0 | 23 | |
| GSE43470 | ASB3 | 51130 | 5 | 0 | 38 | |
| GSE46452 | ASB3 | 51130 | 2 | 4 | 53 | |
| GSE47630 | ASB3 | 51130 | 8 | 0 | 32 | |
| GSE54993 | ASB3 | 51130 | 0 | 5 | 65 | |
| GSE54994 | ASB3 | 51130 | 12 | 0 | 41 | |
| GSE60625 | ASB3 | 51130 | 0 | 3 | 8 | |
| GSE74703 | ASB3 | 51130 | 5 | 0 | 31 | |
| GSE74704 | ASB3 | 51130 | 8 | 0 | 12 | |
| TCGA | ASB3 | 51130 | 36 | 2 | 58 |
Total number of gains: 91; Total number of losses: 14; Total Number of normals: 383.
Somatic mutations of ASB3:
Generating mutation plots.
Highly correlated genes for ASB3:
Showing top 20/56 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ASB3 | ALG11 | 0.915437 | 3 | 0 | 3 |
| ASB3 | REV3L | 0.910145 | 3 | 0 | 3 |
| ASB3 | MIPOL1 | 0.892879 | 3 | 0 | 3 |
| ASB3 | RLIM | 0.854276 | 3 | 0 | 3 |
| ASB3 | ZNF445 | 0.847697 | 3 | 0 | 3 |
| ASB3 | SLC38A6 | 0.824146 | 3 | 0 | 3 |
| ASB3 | SZT2 | 0.816345 | 3 | 0 | 3 |
| ASB3 | NUDT19 | 0.808179 | 3 | 0 | 3 |
| ASB3 | NQO2 | 0.791405 | 3 | 0 | 3 |
| ASB3 | FAM111B | 0.77766 | 3 | 0 | 3 |
| ASB3 | LEPROTL1 | 0.775697 | 3 | 0 | 3 |
| ASB3 | GRIA4 | 0.761244 | 3 | 0 | 3 |
| ASB3 | SMAD2 | 0.758228 | 3 | 0 | 3 |
| ASB3 | NRSN2 | 0.741617 | 3 | 0 | 3 |
| ASB3 | CCDC144A | 0.727067 | 3 | 0 | 3 |
| ASB3 | KRTAP12-2 | 0.726546 | 3 | 0 | 3 |
| ASB3 | OR2T33 | 0.720043 | 3 | 0 | 3 |
| ASB3 | TNFRSF10C | 0.718676 | 3 | 0 | 3 |
| ASB3 | CCDC154 | 0.711205 | 3 | 0 | 3 |
| ASB3 | SLC26A5 | 0.710615 | 3 | 0 | 3 |
For details and further investigation, click here