| Full name: ATPase H+ transporting accessory protein 2 | Alias Symbol: M8-9|APT6M8-9|ATP6M8-9|PRR|RENR | ||
| Type: protein-coding gene | Cytoband: Xp11.4 | ||
| Entrez ID: 10159 | HGNC ID: HGNC:18305 | Ensembl Gene: ENSG00000182220 | OMIM ID: 300556 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of ATP6AP2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ATP6AP2 | 10159 | 201443_s_at | 0.0814 | 0.8706 | |
| GSE20347 | ATP6AP2 | 10159 | 201443_s_at | -0.1975 | 0.1703 | |
| GSE23400 | ATP6AP2 | 10159 | 201443_s_at | -0.0070 | 0.9447 | |
| GSE26886 | ATP6AP2 | 10159 | 201443_s_at | -0.2668 | 0.1475 | |
| GSE29001 | ATP6AP2 | 10159 | 201443_s_at | -0.0364 | 0.9110 | |
| GSE38129 | ATP6AP2 | 10159 | 201443_s_at | -0.0838 | 0.5109 | |
| GSE45670 | ATP6AP2 | 10159 | 201443_s_at | 0.0401 | 0.8590 | |
| GSE53622 | ATP6AP2 | 10159 | 49491 | 0.0830 | 0.2117 | |
| GSE53624 | ATP6AP2 | 10159 | 96741 | 0.0195 | 0.6951 | |
| GSE63941 | ATP6AP2 | 10159 | 201443_s_at | -1.4157 | 0.0082 | |
| GSE77861 | ATP6AP2 | 10159 | 201443_s_at | -0.2084 | 0.3356 | |
| GSE97050 | ATP6AP2 | 10159 | A_23_P11353 | 0.3633 | 0.2203 | |
| SRP007169 | ATP6AP2 | 10159 | RNAseq | -0.6791 | 0.0469 | |
| SRP008496 | ATP6AP2 | 10159 | RNAseq | -0.4566 | 0.0799 | |
| SRP064894 | ATP6AP2 | 10159 | RNAseq | 0.4477 | 0.0033 | |
| SRP133303 | ATP6AP2 | 10159 | RNAseq | 0.3295 | 0.0812 | |
| SRP159526 | ATP6AP2 | 10159 | RNAseq | 0.1170 | 0.5176 | |
| SRP193095 | ATP6AP2 | 10159 | RNAseq | 0.0059 | 0.9646 | |
| SRP219564 | ATP6AP2 | 10159 | RNAseq | 0.2499 | 0.5313 | |
| TCGA | ATP6AP2 | 10159 | RNAseq | 0.0886 | 0.0476 |
Upregulated datasets: 0; Downregulated datasets: 1.
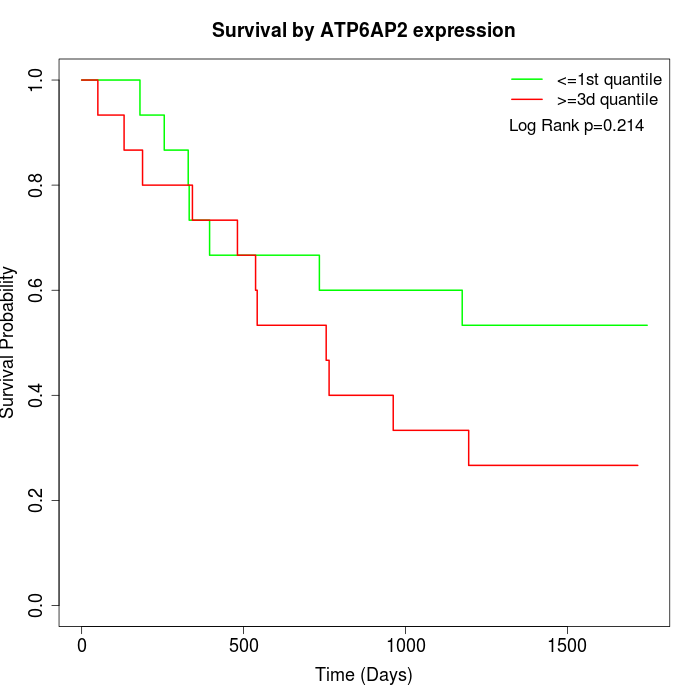
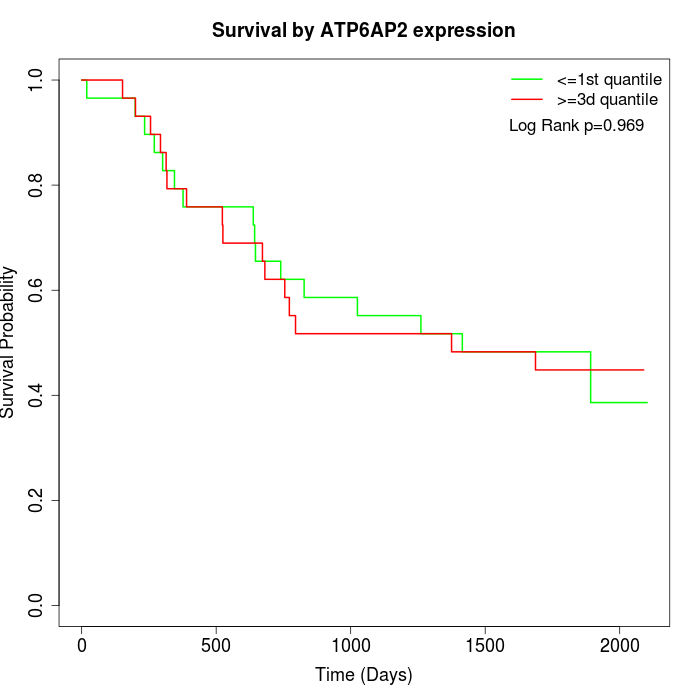
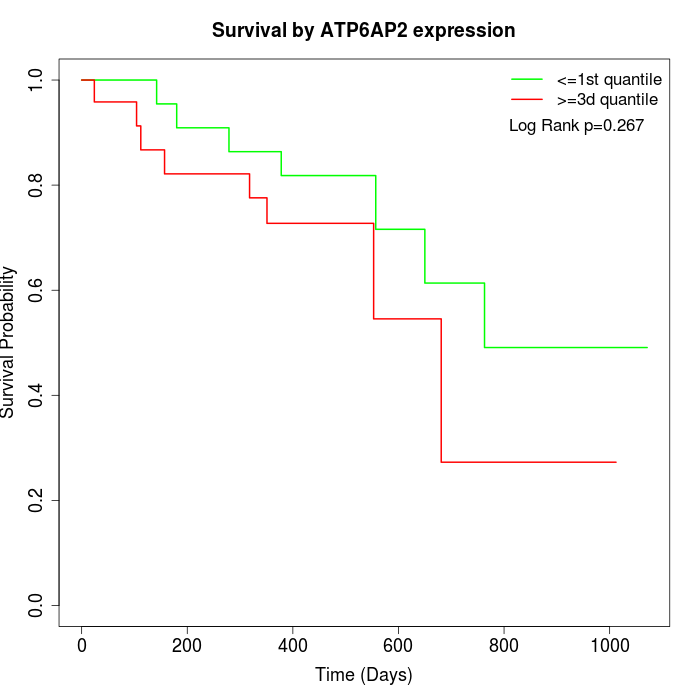
Survival by ATP6AP2 expression:
Note: Click image to view full size file.
Copy number change of ATP6AP2:
No record found for this gene.
Somatic mutations of ATP6AP2:
Generating mutation plots.
Highly correlated genes for ATP6AP2:
Showing top 20/46 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ATP6AP2 | ZNF777 | 0.738785 | 3 | 0 | 3 |
| ATP6AP2 | GIT2 | 0.710216 | 3 | 0 | 3 |
| ATP6AP2 | PHF10 | 0.642861 | 4 | 0 | 3 |
| ATP6AP2 | MAP3K4 | 0.642552 | 4 | 0 | 4 |
| ATP6AP2 | FUNDC1 | 0.639791 | 6 | 0 | 6 |
| ATP6AP2 | ADPRHL2 | 0.63381 | 4 | 0 | 3 |
| ATP6AP2 | RWDD3 | 0.632502 | 4 | 0 | 3 |
| ATP6AP2 | EEF1A1 | 0.62369 | 3 | 0 | 3 |
| ATP6AP2 | ZNF230 | 0.60931 | 3 | 0 | 3 |
| ATP6AP2 | ARFGEF1 | 0.60027 | 6 | 0 | 4 |
| ATP6AP2 | WDR60 | 0.592427 | 3 | 0 | 3 |
| ATP6AP2 | ZNF500 | 0.59003 | 5 | 0 | 4 |
| ATP6AP2 | PDZRN3 | 0.586534 | 5 | 0 | 3 |
| ATP6AP2 | MFSD1 | 0.583269 | 7 | 0 | 5 |
| ATP6AP2 | CCDC96 | 0.582671 | 5 | 0 | 3 |
| ATP6AP2 | PAFAH1B1 | 0.575553 | 4 | 0 | 3 |
| ATP6AP2 | CYP2E1 | 0.571686 | 3 | 0 | 3 |
| ATP6AP2 | TMA7 | 0.568611 | 5 | 0 | 3 |
| ATP6AP2 | ERMP1 | 0.568018 | 3 | 0 | 3 |
| ATP6AP2 | PAN3-AS1 | 0.567824 | 3 | 0 | 3 |
For details and further investigation, click here