| Full name: eukaryotic translation elongation factor 1 alpha 1 | Alias Symbol: EE1A1 | ||
| Type: protein-coding gene | Cytoband: 6q13 | ||
| Entrez ID: 1915 | HGNC ID: HGNC:3189 | Ensembl Gene: ENSG00000156508 | OMIM ID: 130590 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
EEF1A1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa05134 | Legionellosis |
Expression of EEF1A1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | EEF1A1 | 1915 | 204892_x_at | -0.1140 | 0.5729 | |
| GSE20347 | EEF1A1 | 1915 | 213477_x_at | 0.0380 | 0.4078 | |
| GSE23400 | EEF1A1 | 1915 | 204892_x_at | -0.0425 | 0.2810 | |
| GSE26886 | EEF1A1 | 1915 | 204892_x_at | -0.4193 | 0.0005 | |
| GSE29001 | EEF1A1 | 1915 | 213477_x_at | -0.1258 | 0.4030 | |
| GSE38129 | EEF1A1 | 1915 | 204892_x_at | -0.0111 | 0.8707 | |
| GSE45670 | EEF1A1 | 1915 | 204892_x_at | -0.1636 | 0.0016 | |
| GSE53622 | EEF1A1 | 1915 | 64190 | -0.2773 | 0.0002 | |
| GSE53624 | EEF1A1 | 1915 | 65985 | -0.1467 | 0.0159 | |
| GSE63941 | EEF1A1 | 1915 | 204892_x_at | 0.0003 | 0.9982 | |
| GSE77861 | EEF1A1 | 1915 | 204892_x_at | -0.0806 | 0.5397 | |
| GSE97050 | EEF1A1 | 1915 | A_32_P47701 | -0.2518 | 0.3261 | |
| SRP007169 | EEF1A1 | 1915 | RNAseq | -0.7010 | 0.0216 | |
| SRP008496 | EEF1A1 | 1915 | RNAseq | -0.6184 | 0.0083 | |
| SRP064894 | EEF1A1 | 1915 | RNAseq | -0.7485 | 0.0001 | |
| SRP133303 | EEF1A1 | 1915 | RNAseq | -0.7717 | 0.0000 | |
| SRP159526 | EEF1A1 | 1915 | RNAseq | -0.8580 | 0.0099 | |
| SRP193095 | EEF1A1 | 1915 | RNAseq | -1.1220 | 0.0000 | |
| SRP219564 | EEF1A1 | 1915 | RNAseq | -0.7910 | 0.0011 | |
| TCGA | EEF1A1 | 1915 | RNAseq | -0.1054 | 0.0014 |
Upregulated datasets: 0; Downregulated datasets: 1.
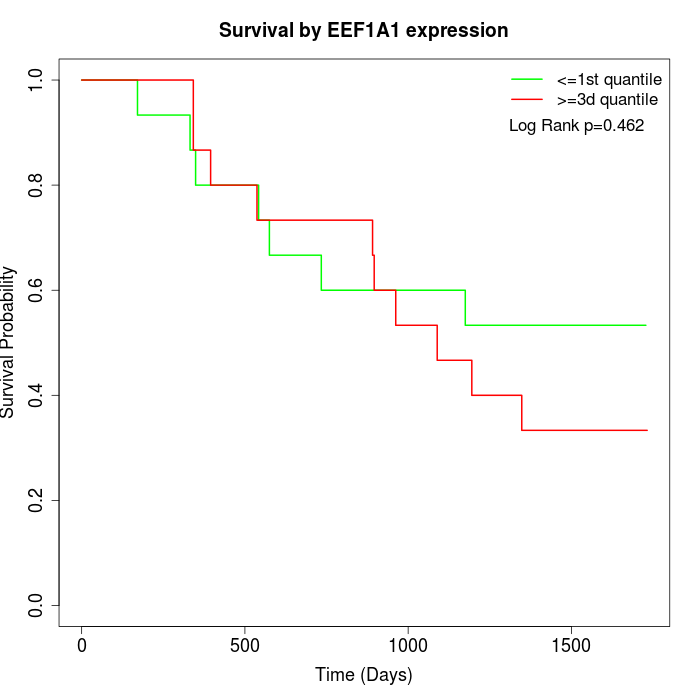
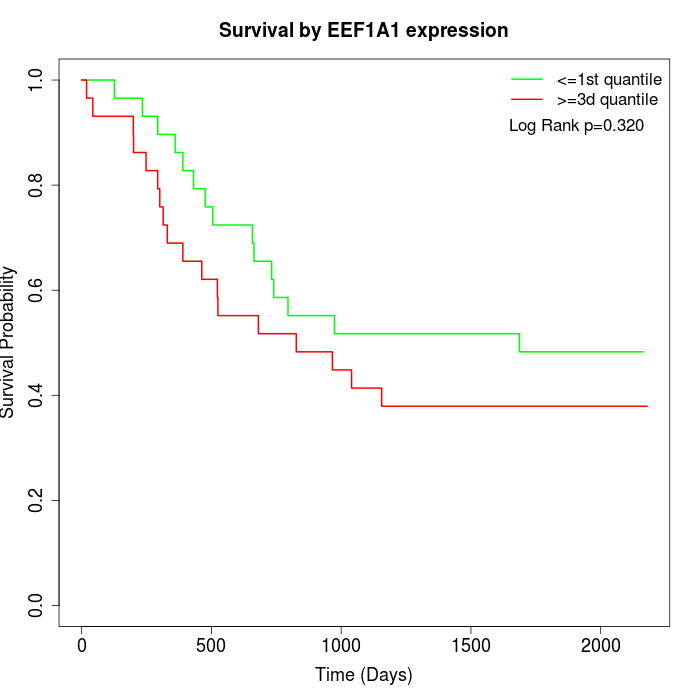
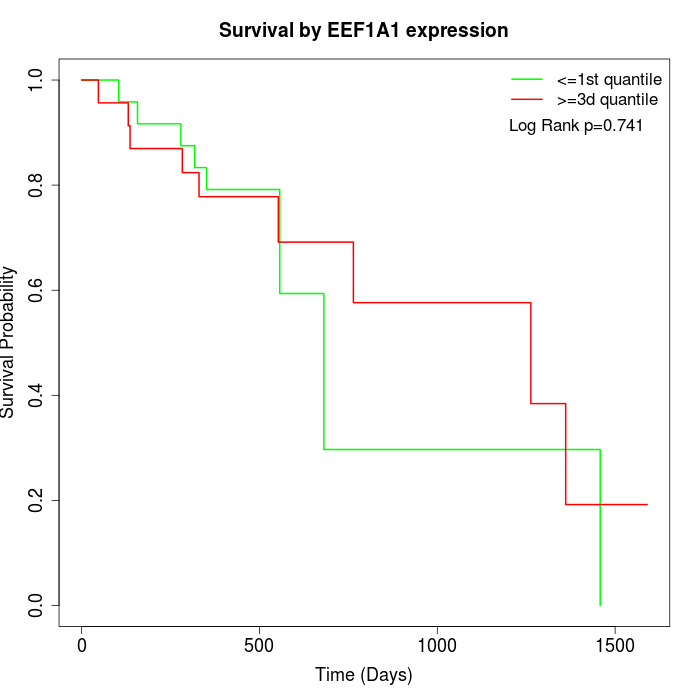
Survival by EEF1A1 expression:
Note: Click image to view full size file.
Copy number change of EEF1A1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | EEF1A1 | 1915 | 1 | 3 | 26 | |
| GSE20123 | EEF1A1 | 1915 | 1 | 2 | 27 | |
| GSE43470 | EEF1A1 | 1915 | 3 | 0 | 40 | |
| GSE46452 | EEF1A1 | 1915 | 2 | 11 | 46 | |
| GSE47630 | EEF1A1 | 1915 | 8 | 4 | 28 | |
| GSE54993 | EEF1A1 | 1915 | 3 | 2 | 65 | |
| GSE54994 | EEF1A1 | 1915 | 8 | 9 | 36 | |
| GSE60625 | EEF1A1 | 1915 | 0 | 1 | 10 | |
| GSE74703 | EEF1A1 | 1915 | 3 | 0 | 33 | |
| GSE74704 | EEF1A1 | 1915 | 0 | 1 | 19 | |
| TCGA | EEF1A1 | 1915 | 10 | 21 | 65 |
Total number of gains: 39; Total number of losses: 54; Total Number of normals: 395.
Somatic mutations of EEF1A1:
Generating mutation plots.
Highly correlated genes for EEF1A1:
Showing top 20/239 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| EEF1A1 | RPL3 | 0.715446 | 8 | 0 | 7 |
| EEF1A1 | RPS28 | 0.698473 | 3 | 0 | 3 |
| EEF1A1 | RPS9 | 0.695167 | 8 | 0 | 7 |
| EEF1A1 | TMEM167B | 0.694161 | 4 | 0 | 4 |
| EEF1A1 | HBP1 | 0.689428 | 3 | 0 | 3 |
| EEF1A1 | ACO2 | 0.684512 | 4 | 0 | 3 |
| EEF1A1 | TMEM50A | 0.682387 | 4 | 0 | 4 |
| EEF1A1 | BCDIN3D | 0.681848 | 3 | 0 | 3 |
| EEF1A1 | RPL15 | 0.677501 | 8 | 0 | 7 |
| EEF1A1 | CARF | 0.675845 | 3 | 0 | 3 |
| EEF1A1 | PPP2CB | 0.673974 | 3 | 0 | 3 |
| EEF1A1 | FBXL5 | 0.668749 | 5 | 0 | 4 |
| EEF1A1 | FBXL3 | 0.654242 | 4 | 0 | 4 |
| EEF1A1 | ATF6 | 0.650388 | 3 | 0 | 3 |
| EEF1A1 | TRAK2 | 0.649652 | 5 | 0 | 3 |
| EEF1A1 | RPS5 | 0.649122 | 7 | 0 | 7 |
| EEF1A1 | VAMP5 | 0.648886 | 3 | 0 | 3 |
| EEF1A1 | RNF125 | 0.647448 | 3 | 0 | 3 |
| EEF1A1 | RPL10A | 0.643588 | 9 | 0 | 9 |
| EEF1A1 | ZMYND11 | 0.638863 | 4 | 0 | 3 |
For details and further investigation, click here