| Full name: GIT ArfGAP 2 | Alias Symbol: KIAA0148|PKL | ||
| Type: protein-coding gene | Cytoband: 12q24.11 | ||
| Entrez ID: 9815 | HGNC ID: HGNC:4273 | Ensembl Gene: ENSG00000139436 | OMIM ID: 608564 |
| Drug and gene relationship at DGIdb | |||
Expression of GIT2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GIT2 | 9815 | 225558_at | -0.0320 | 0.9479 | |
| GSE20347 | GIT2 | 9815 | 209876_at | 0.0029 | 0.9781 | |
| GSE23400 | GIT2 | 9815 | 204982_at | -0.0661 | 0.0352 | |
| GSE26886 | GIT2 | 9815 | 225558_at | -0.3986 | 0.1115 | |
| GSE29001 | GIT2 | 9815 | 204982_at | 0.1946 | 0.2956 | |
| GSE38129 | GIT2 | 9815 | 204982_at | 0.0872 | 0.6374 | |
| GSE45670 | GIT2 | 9815 | 225558_at | -0.1169 | 0.4457 | |
| GSE53622 | GIT2 | 9815 | 62883 | -0.1786 | 0.0134 | |
| GSE53624 | GIT2 | 9815 | 62883 | -0.1280 | 0.0344 | |
| GSE63941 | GIT2 | 9815 | 225558_at | -1.4776 | 0.0015 | |
| GSE77861 | GIT2 | 9815 | 225558_at | 0.1016 | 0.6473 | |
| GSE97050 | GIT2 | 9815 | A_23_P204436 | 0.0731 | 0.8171 | |
| SRP007169 | GIT2 | 9815 | RNAseq | 0.8771 | 0.0673 | |
| SRP008496 | GIT2 | 9815 | RNAseq | 0.8795 | 0.0026 | |
| SRP064894 | GIT2 | 9815 | RNAseq | 0.3253 | 0.0096 | |
| SRP133303 | GIT2 | 9815 | RNAseq | -0.0272 | 0.8338 | |
| SRP159526 | GIT2 | 9815 | RNAseq | -0.0185 | 0.9220 | |
| SRP193095 | GIT2 | 9815 | RNAseq | 0.0688 | 0.4664 | |
| SRP219564 | GIT2 | 9815 | RNAseq | 0.1992 | 0.3350 | |
| TCGA | GIT2 | 9815 | RNAseq | -0.0828 | 0.0790 |
Upregulated datasets: 0; Downregulated datasets: 1.
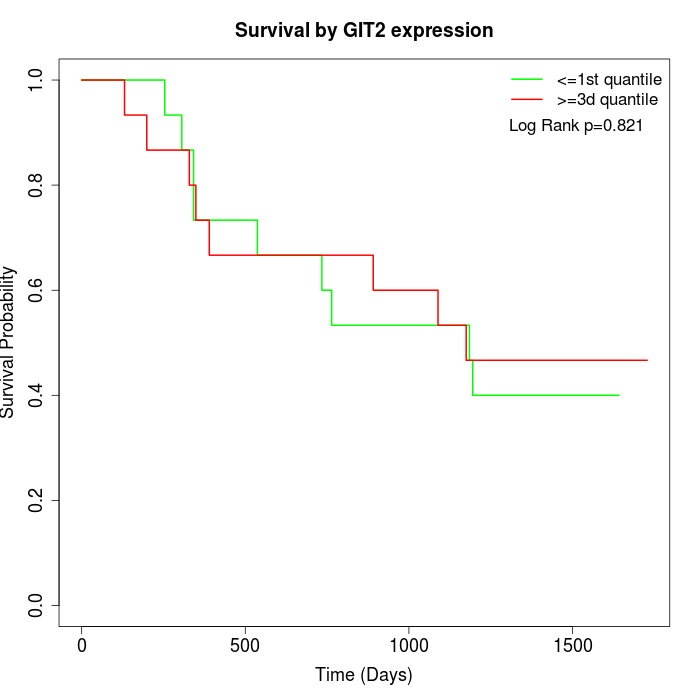
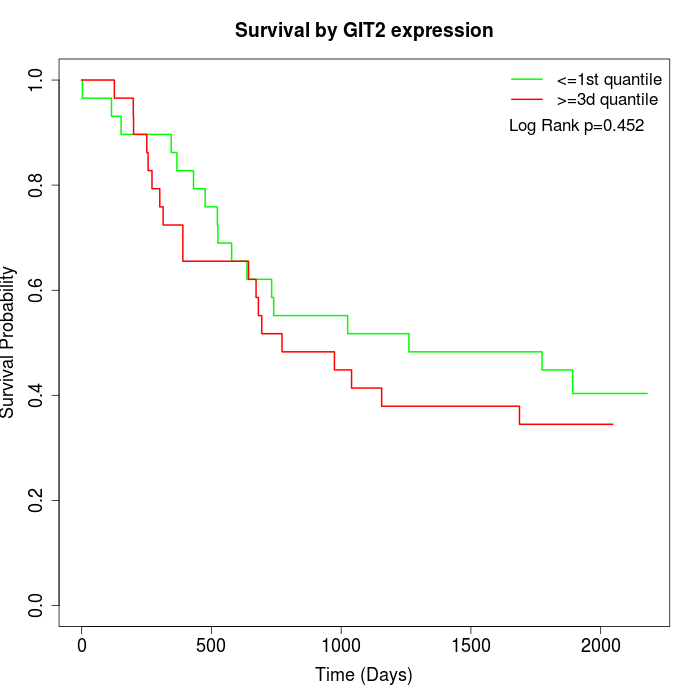
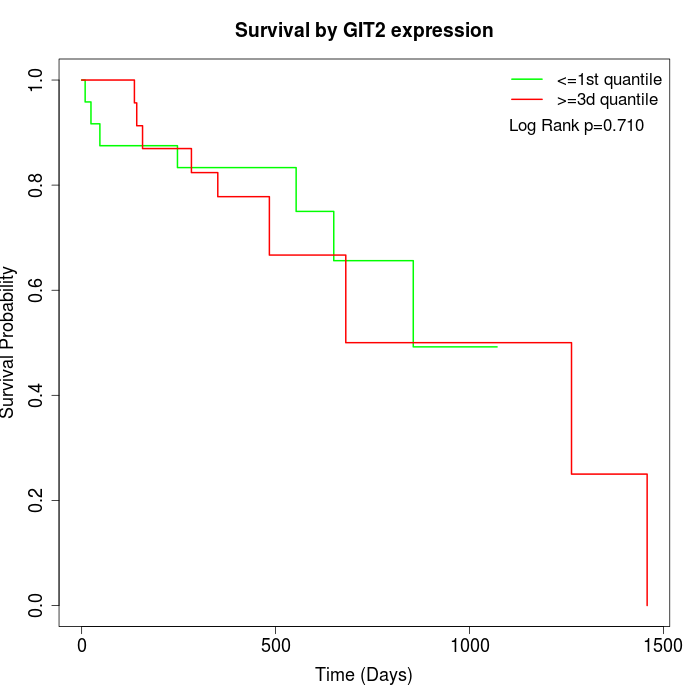
Survival by GIT2 expression:
Note: Click image to view full size file.
Copy number change of GIT2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GIT2 | 9815 | 5 | 3 | 22 | |
| GSE20123 | GIT2 | 9815 | 5 | 3 | 22 | |
| GSE43470 | GIT2 | 9815 | 2 | 1 | 40 | |
| GSE46452 | GIT2 | 9815 | 9 | 1 | 49 | |
| GSE47630 | GIT2 | 9815 | 9 | 1 | 30 | |
| GSE54993 | GIT2 | 9815 | 0 | 5 | 65 | |
| GSE54994 | GIT2 | 9815 | 4 | 3 | 46 | |
| GSE60625 | GIT2 | 9815 | 0 | 0 | 11 | |
| GSE74703 | GIT2 | 9815 | 2 | 0 | 34 | |
| GSE74704 | GIT2 | 9815 | 2 | 2 | 16 | |
| TCGA | GIT2 | 9815 | 22 | 10 | 64 |
Total number of gains: 60; Total number of losses: 29; Total Number of normals: 399.
Somatic mutations of GIT2:
Generating mutation plots.
Highly correlated genes for GIT2:
Showing top 20/66 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GIT2 | ATP6AP2 | 0.710216 | 3 | 0 | 3 |
| GIT2 | FAM110B | 0.695558 | 4 | 0 | 3 |
| GIT2 | ZNF337 | 0.693265 | 3 | 0 | 3 |
| GIT2 | EMILIN1 | 0.666613 | 4 | 0 | 4 |
| GIT2 | PRAF2 | 0.662656 | 3 | 0 | 3 |
| GIT2 | ENTPD1 | 0.634168 | 5 | 0 | 4 |
| GIT2 | CYB561D2 | 0.630819 | 4 | 0 | 3 |
| GIT2 | SFRP1 | 0.628707 | 3 | 0 | 3 |
| GIT2 | EFR3A | 0.627951 | 3 | 0 | 3 |
| GIT2 | SCRN2 | 0.623702 | 3 | 0 | 3 |
| GIT2 | ATP8B4 | 0.609988 | 6 | 0 | 5 |
| GIT2 | DOCK2 | 0.608224 | 7 | 0 | 6 |
| GIT2 | TMCO3 | 0.607293 | 5 | 0 | 3 |
| GIT2 | CARD8 | 0.607147 | 6 | 0 | 3 |
| GIT2 | NICN1 | 0.606886 | 4 | 0 | 3 |
| GIT2 | ZNF197 | 0.599432 | 4 | 0 | 3 |
| GIT2 | MANBA | 0.596879 | 4 | 0 | 3 |
| GIT2 | GREM1 | 0.594692 | 4 | 0 | 3 |
| GIT2 | LY6G5C | 0.589236 | 5 | 0 | 3 |
| GIT2 | VCAM1 | 0.588591 | 4 | 0 | 3 |
For details and further investigation, click here