| Full name: ATPase H+ transporting V0 subunit a4 | Alias Symbol: RDRTA2|VPP2|RTADR|a4|Vph1|Stv1 | ||
| Type: protein-coding gene | Cytoband: 7q34 | ||
| Entrez ID: 50617 | HGNC ID: HGNC:866 | Ensembl Gene: ENSG00000105929 | OMIM ID: 605239 |
| Drug and gene relationship at DGIdb | |||
ATP6V0A4 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa05110 | Vibrio cholerae infection | |
| hsa05120 | Epithelial cell signaling in Helicobacter pylori infection | |
| hsa05152 | Tuberculosis |
Expression of ATP6V0A4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ATP6V0A4 | 50617 | 220197_at | -1.6478 | 0.0965 | |
| GSE20347 | ATP6V0A4 | 50617 | 220197_at | -0.8879 | 0.0000 | |
| GSE23400 | ATP6V0A4 | 50617 | 220197_at | -0.5326 | 0.0000 | |
| GSE26886 | ATP6V0A4 | 50617 | 220197_at | -2.7590 | 0.0000 | |
| GSE29001 | ATP6V0A4 | 50617 | 220197_at | -1.2849 | 0.0045 | |
| GSE38129 | ATP6V0A4 | 50617 | 220197_at | -0.8083 | 0.0002 | |
| GSE45670 | ATP6V0A4 | 50617 | 220197_at | 0.1704 | 0.8053 | |
| GSE53622 | ATP6V0A4 | 50617 | 16242 | -2.3165 | 0.0000 | |
| GSE53624 | ATP6V0A4 | 50617 | 16242 | -2.9027 | 0.0000 | |
| GSE63941 | ATP6V0A4 | 50617 | 220197_at | 0.4532 | 0.4050 | |
| GSE77861 | ATP6V0A4 | 50617 | 220197_at | -0.7067 | 0.0027 | |
| GSE97050 | ATP6V0A4 | 50617 | A_23_P215111 | 0.1627 | 0.5551 | |
| SRP007169 | ATP6V0A4 | 50617 | RNAseq | -3.4083 | 0.0000 | |
| SRP008496 | ATP6V0A4 | 50617 | RNAseq | -3.9734 | 0.0000 | |
| SRP064894 | ATP6V0A4 | 50617 | RNAseq | -3.2613 | 0.0000 | |
| SRP133303 | ATP6V0A4 | 50617 | RNAseq | -2.4337 | 0.0000 | |
| SRP159526 | ATP6V0A4 | 50617 | RNAseq | -3.8801 | 0.0000 | |
| TCGA | ATP6V0A4 | 50617 | RNAseq | 1.1076 | 0.1141 |
Upregulated datasets: 0; Downregulated datasets: 9.
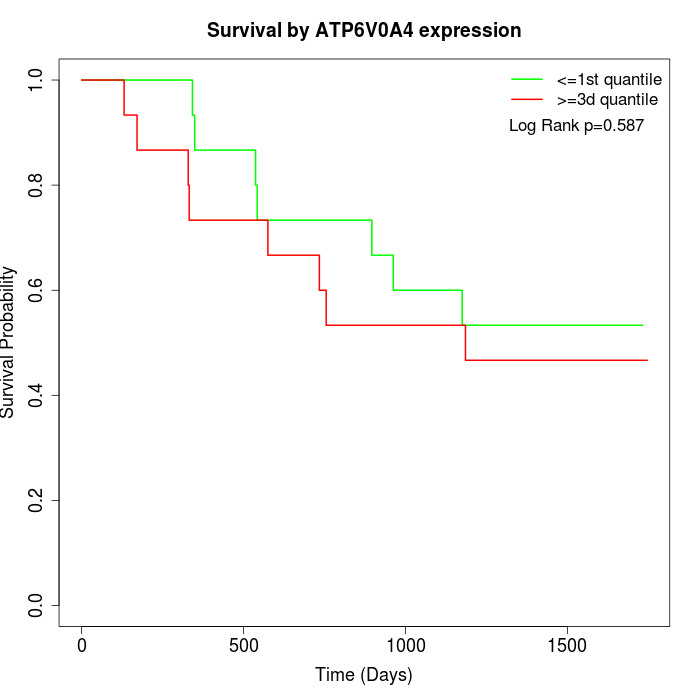
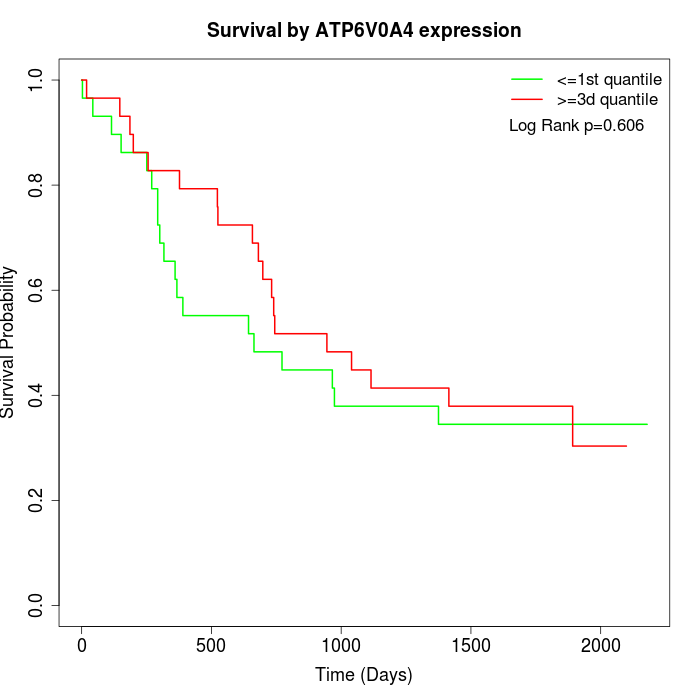
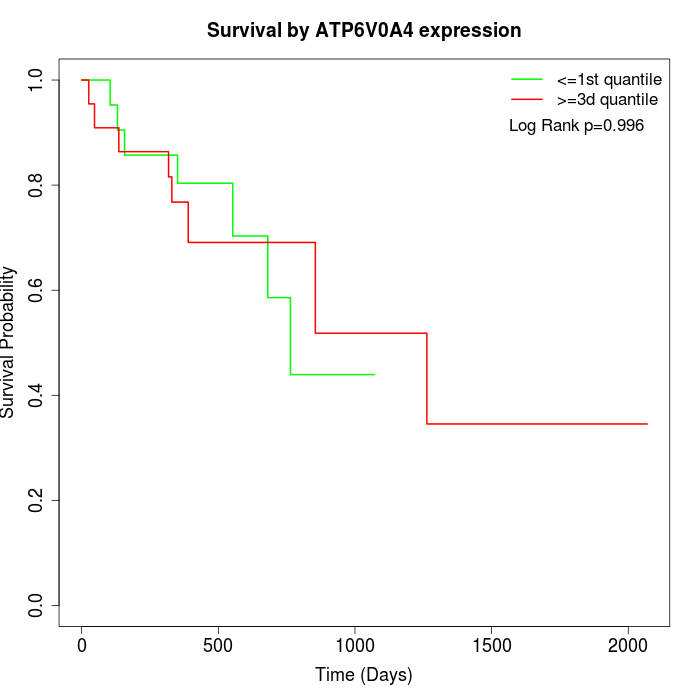
Survival by ATP6V0A4 expression:
Note: Click image to view full size file.
Copy number change of ATP6V0A4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ATP6V0A4 | 50617 | 4 | 2 | 24 | |
| GSE20123 | ATP6V0A4 | 50617 | 4 | 2 | 24 | |
| GSE43470 | ATP6V0A4 | 50617 | 2 | 3 | 38 | |
| GSE46452 | ATP6V0A4 | 50617 | 7 | 2 | 50 | |
| GSE47630 | ATP6V0A4 | 50617 | 7 | 4 | 29 | |
| GSE54993 | ATP6V0A4 | 50617 | 3 | 5 | 62 | |
| GSE54994 | ATP6V0A4 | 50617 | 5 | 8 | 40 | |
| GSE60625 | ATP6V0A4 | 50617 | 0 | 0 | 11 | |
| GSE74703 | ATP6V0A4 | 50617 | 2 | 3 | 31 | |
| GSE74704 | ATP6V0A4 | 50617 | 1 | 2 | 17 | |
| TCGA | ATP6V0A4 | 50617 | 28 | 23 | 45 |
Total number of gains: 63; Total number of losses: 54; Total Number of normals: 371.
Somatic mutations of ATP6V0A4:
Generating mutation plots.
Highly correlated genes for ATP6V0A4:
Showing top 20/881 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ATP6V0A4 | PRSS27 | 0.827595 | 7 | 0 | 7 |
| ATP6V0A4 | CAPN14 | 0.815388 | 6 | 0 | 6 |
| ATP6V0A4 | NCCRP1 | 0.810963 | 3 | 0 | 3 |
| ATP6V0A4 | ATP13A4 | 0.806963 | 6 | 0 | 6 |
| ATP6V0A4 | C3orf20 | 0.803208 | 3 | 0 | 3 |
| ATP6V0A4 | IL36A | 0.801874 | 11 | 0 | 11 |
| ATP6V0A4 | TRNP1 | 0.794363 | 6 | 0 | 6 |
| ATP6V0A4 | KRT78 | 0.787706 | 6 | 0 | 6 |
| ATP6V0A4 | FLG-AS1 | 0.778993 | 6 | 0 | 6 |
| ATP6V0A4 | VSIG10L | 0.777108 | 7 | 0 | 7 |
| ATP6V0A4 | FAM163B | 0.776991 | 3 | 0 | 3 |
| ATP6V0A4 | C15orf48 | 0.772386 | 7 | 0 | 7 |
| ATP6V0A4 | ANXA9 | 0.771309 | 11 | 0 | 10 |
| ATP6V0A4 | GCHFR | 0.766216 | 9 | 0 | 9 |
| ATP6V0A4 | CEACAM7 | 0.765873 | 11 | 0 | 11 |
| ATP6V0A4 | SCNN1B | 0.765754 | 11 | 0 | 11 |
| ATP6V0A4 | MAB21L3 | 0.76497 | 6 | 0 | 6 |
| ATP6V0A4 | TMEM45B | 0.763108 | 6 | 0 | 6 |
| ATP6V0A4 | SLC13A4 | 0.760726 | 12 | 0 | 11 |
| ATP6V0A4 | KLB | 0.760396 | 6 | 0 | 6 |
For details and further investigation, click here