| Full name: ATPase H+ transporting V1 subunit H | Alias Symbol: CGI-11|SFD|VMA13|SFDalpha|SFDbeta | ||
| Type: protein-coding gene | Cytoband: 8q11.23 | ||
| Entrez ID: 51606 | HGNC ID: HGNC:18303 | Ensembl Gene: ENSG00000047249 | OMIM ID: 608861 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
ATP6V1H involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04150 | mTOR signaling pathway | |
| hsa05110 | Vibrio cholerae infection | |
| hsa05120 | Epithelial cell signaling in Helicobacter pylori infection | |
| hsa05152 | Tuberculosis |
Expression of ATP6V1H:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ATP6V1H | 51606 | 221504_s_at | 0.1461 | 0.7169 | |
| GSE20347 | ATP6V1H | 51606 | 221504_s_at | -0.0708 | 0.7116 | |
| GSE23400 | ATP6V1H | 51606 | 221504_s_at | -0.0639 | 0.3418 | |
| GSE26886 | ATP6V1H | 51606 | 221504_s_at | -0.8473 | 0.0014 | |
| GSE29001 | ATP6V1H | 51606 | 221504_s_at | -0.0699 | 0.8763 | |
| GSE38129 | ATP6V1H | 51606 | 221504_s_at | -0.1050 | 0.4867 | |
| GSE45670 | ATP6V1H | 51606 | 221504_s_at | -0.1617 | 0.1146 | |
| GSE53622 | ATP6V1H | 51606 | 7012 | -0.1867 | 0.0062 | |
| GSE53624 | ATP6V1H | 51606 | 7012 | -0.4453 | 0.0000 | |
| GSE63941 | ATP6V1H | 51606 | 221504_s_at | -1.0095 | 0.0156 | |
| GSE77861 | ATP6V1H | 51606 | 221504_s_at | -0.1967 | 0.4075 | |
| GSE97050 | ATP6V1H | 51606 | A_24_P363679 | -0.0595 | 0.8543 | |
| SRP007169 | ATP6V1H | 51606 | RNAseq | -1.3504 | 0.0002 | |
| SRP008496 | ATP6V1H | 51606 | RNAseq | -0.8800 | 0.0002 | |
| SRP064894 | ATP6V1H | 51606 | RNAseq | -0.0459 | 0.7971 | |
| SRP133303 | ATP6V1H | 51606 | RNAseq | -0.1434 | 0.3673 | |
| SRP159526 | ATP6V1H | 51606 | RNAseq | -0.4624 | 0.0906 | |
| SRP193095 | ATP6V1H | 51606 | RNAseq | -0.3572 | 0.0075 | |
| SRP219564 | ATP6V1H | 51606 | RNAseq | -0.1897 | 0.5183 | |
| TCGA | ATP6V1H | 51606 | RNAseq | -0.1166 | 0.0130 |
Upregulated datasets: 0; Downregulated datasets: 2.
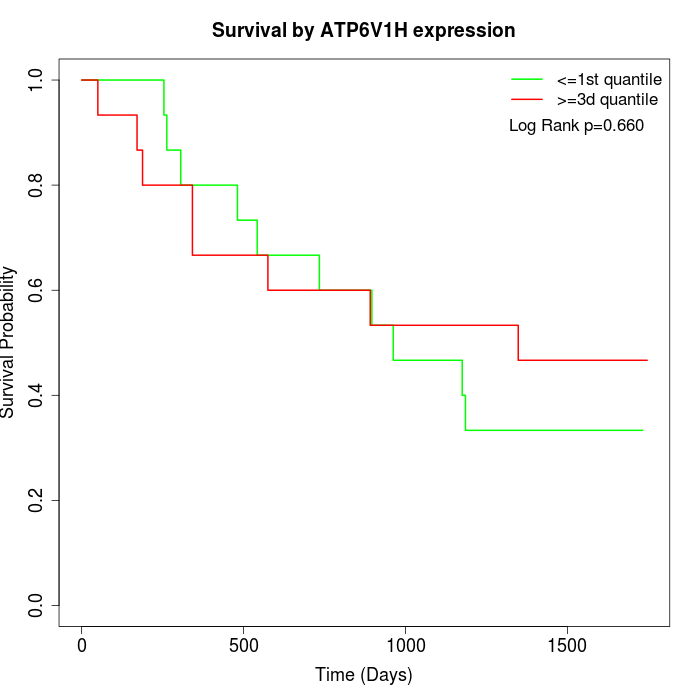
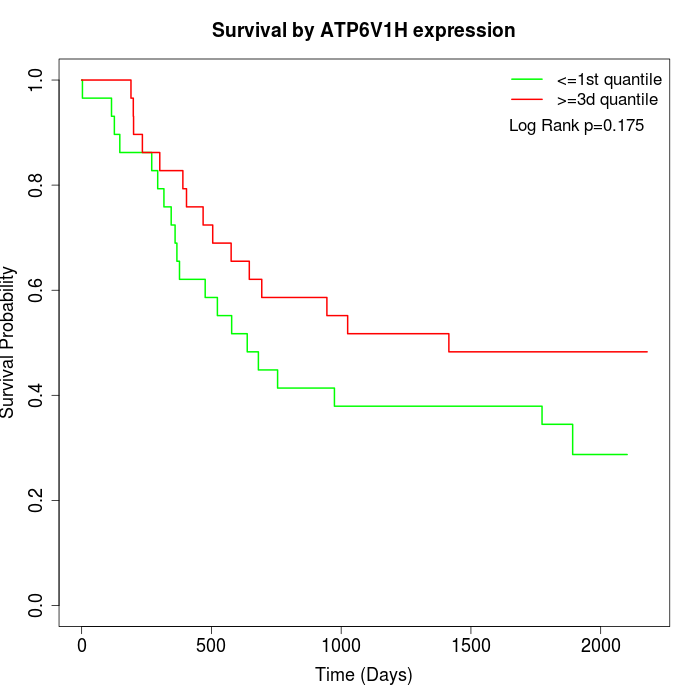
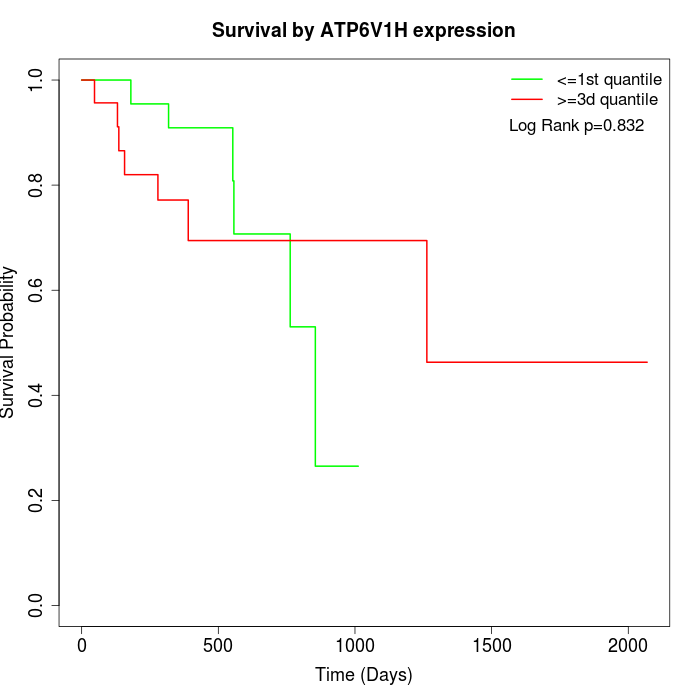
Survival by ATP6V1H expression:
Note: Click image to view full size file.
Copy number change of ATP6V1H:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ATP6V1H | 51606 | 15 | 2 | 13 | |
| GSE20123 | ATP6V1H | 51606 | 15 | 2 | 13 | |
| GSE43470 | ATP6V1H | 51606 | 14 | 3 | 26 | |
| GSE46452 | ATP6V1H | 51606 | 20 | 3 | 36 | |
| GSE47630 | ATP6V1H | 51606 | 23 | 1 | 16 | |
| GSE54993 | ATP6V1H | 51606 | 1 | 16 | 53 | |
| GSE54994 | ATP6V1H | 51606 | 28 | 0 | 25 | |
| GSE60625 | ATP6V1H | 51606 | 0 | 4 | 7 | |
| GSE74703 | ATP6V1H | 51606 | 13 | 2 | 21 | |
| GSE74704 | ATP6V1H | 51606 | 11 | 0 | 9 | |
| TCGA | ATP6V1H | 51606 | 43 | 9 | 44 |
Total number of gains: 183; Total number of losses: 42; Total Number of normals: 263.
Somatic mutations of ATP6V1H:
Generating mutation plots.
Highly correlated genes for ATP6V1H:
Showing top 20/393 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ATP6V1H | RBPJ | 0.809432 | 3 | 0 | 3 |
| ATP6V1H | VPS26B | 0.801102 | 3 | 0 | 3 |
| ATP6V1H | AMZ2 | 0.776331 | 3 | 0 | 3 |
| ATP6V1H | TBRG1 | 0.76776 | 3 | 0 | 3 |
| ATP6V1H | ZNF547 | 0.752835 | 3 | 0 | 3 |
| ATP6V1H | NIPAL3 | 0.750645 | 4 | 0 | 4 |
| ATP6V1H | MRFAP1L1 | 0.743191 | 4 | 0 | 4 |
| ATP6V1H | ZBED6 | 0.737741 | 3 | 0 | 3 |
| ATP6V1H | HNRNPH2 | 0.736466 | 3 | 0 | 3 |
| ATP6V1H | PARD3 | 0.729085 | 3 | 0 | 3 |
| ATP6V1H | CYB5A | 0.727433 | 4 | 0 | 4 |
| ATP6V1H | KLHDC2 | 0.727249 | 3 | 0 | 3 |
| ATP6V1H | COMMD9 | 0.72702 | 3 | 0 | 3 |
| ATP6V1H | RBBP4 | 0.726067 | 3 | 0 | 3 |
| ATP6V1H | HMGN1 | 0.72546 | 3 | 0 | 3 |
| ATP6V1H | BRWD3 | 0.724799 | 3 | 0 | 3 |
| ATP6V1H | PIGG | 0.724771 | 3 | 0 | 3 |
| ATP6V1H | NBPF1 | 0.718052 | 3 | 0 | 3 |
| ATP6V1H | PPIP5K2 | 0.713128 | 4 | 0 | 4 |
| ATP6V1H | UBE2J1 | 0.71139 | 4 | 0 | 4 |
For details and further investigation, click here