| Full name: par-3 family cell polarity regulator | Alias Symbol: PAR3|PARD3A|Bazooka|Baz|ASIP|PPP1R118 | ||
| Type: protein-coding gene | Cytoband: 10p11.22-p11.21 | ||
| Entrez ID: 56288 | HGNC ID: HGNC:16051 | Ensembl Gene: ENSG00000148498 | OMIM ID: 606745 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
PARD3 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04015 | Rap1 signaling pathway | |
| hsa04062 | Chemokine signaling pathway | |
| hsa04520 | Adherens junction | |
| hsa04530 | Tight junction |
Expression of PARD3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PARD3 | 56288 | 221526_x_at | -0.5630 | 0.0479 | |
| GSE20347 | PARD3 | 56288 | 221526_x_at | -0.8049 | 0.0001 | |
| GSE23400 | PARD3 | 56288 | 221526_x_at | -0.7423 | 0.0000 | |
| GSE26886 | PARD3 | 56288 | 210094_s_at | -0.7955 | 0.0140 | |
| GSE29001 | PARD3 | 56288 | 210094_s_at | -0.5411 | 0.1030 | |
| GSE38129 | PARD3 | 56288 | 210094_s_at | -0.6752 | 0.0019 | |
| GSE45670 | PARD3 | 56288 | 210094_s_at | -0.3370 | 0.1735 | |
| GSE53622 | PARD3 | 56288 | 18598 | -0.8141 | 0.0000 | |
| GSE53624 | PARD3 | 56288 | 18598 | -0.7942 | 0.0000 | |
| GSE63941 | PARD3 | 56288 | 210094_s_at | -0.3494 | 0.6661 | |
| GSE77861 | PARD3 | 56288 | 221526_x_at | -0.6096 | 0.0447 | |
| GSE97050 | PARD3 | 56288 | A_33_P3303372 | -0.4777 | 0.1049 | |
| SRP007169 | PARD3 | 56288 | RNAseq | -1.7654 | 0.0000 | |
| SRP008496 | PARD3 | 56288 | RNAseq | -1.0817 | 0.0009 | |
| SRP064894 | PARD3 | 56288 | RNAseq | -1.4927 | 0.0000 | |
| SRP133303 | PARD3 | 56288 | RNAseq | -1.0182 | 0.0038 | |
| SRP159526 | PARD3 | 56288 | RNAseq | -0.7831 | 0.1248 | |
| SRP193095 | PARD3 | 56288 | RNAseq | -1.1238 | 0.0000 | |
| SRP219564 | PARD3 | 56288 | RNAseq | -1.5386 | 0.0001 | |
| TCGA | PARD3 | 56288 | RNAseq | 0.0014 | 0.9899 |
Upregulated datasets: 0; Downregulated datasets: 6.
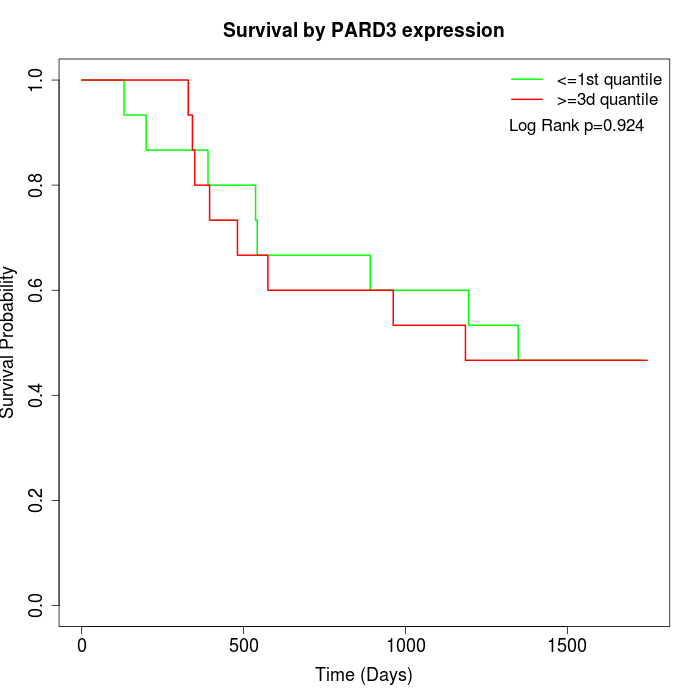
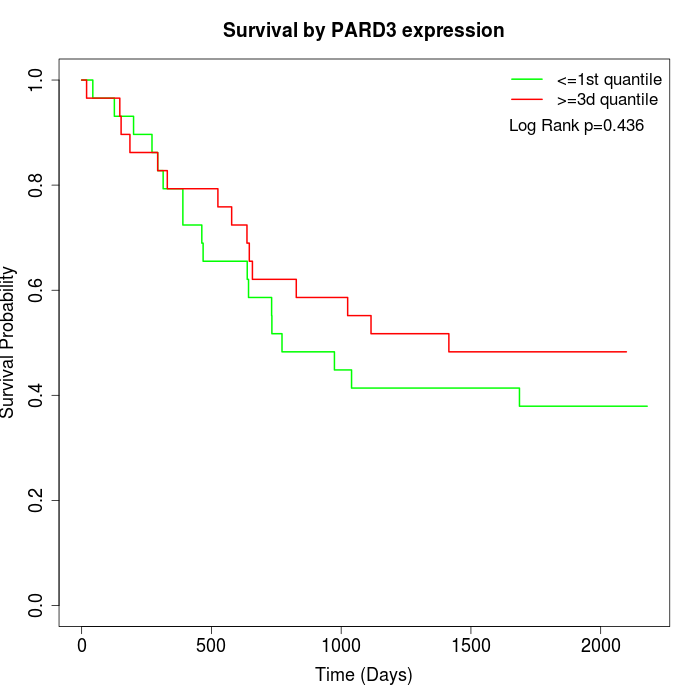
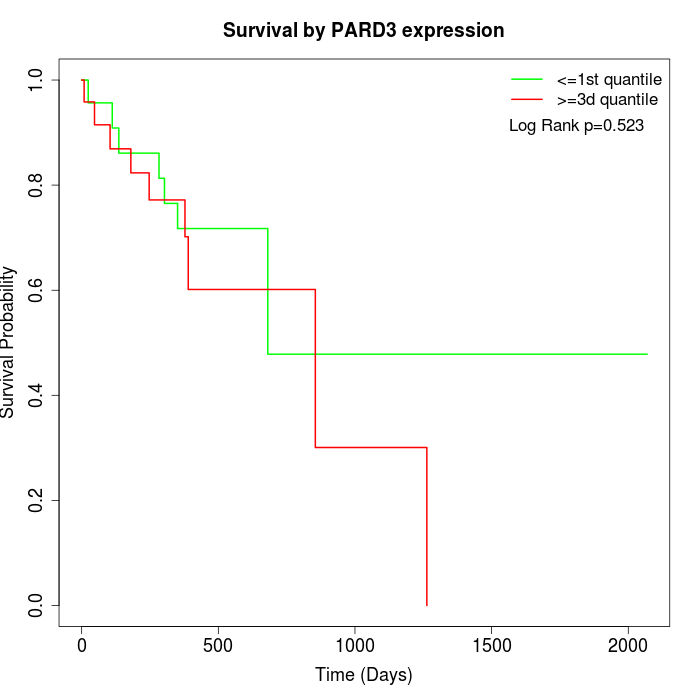
Survival by PARD3 expression:
Note: Click image to view full size file.
Copy number change of PARD3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PARD3 | 56288 | 5 | 7 | 18 | |
| GSE20123 | PARD3 | 56288 | 5 | 6 | 19 | |
| GSE43470 | PARD3 | 56288 | 3 | 7 | 33 | |
| GSE46452 | PARD3 | 56288 | 2 | 13 | 44 | |
| GSE47630 | PARD3 | 56288 | 7 | 13 | 20 | |
| GSE54993 | PARD3 | 56288 | 10 | 0 | 60 | |
| GSE54994 | PARD3 | 56288 | 3 | 14 | 36 | |
| GSE60625 | PARD3 | 56288 | 0 | 0 | 11 | |
| GSE74703 | PARD3 | 56288 | 2 | 5 | 29 | |
| GSE74704 | PARD3 | 56288 | 0 | 5 | 15 | |
| TCGA | PARD3 | 56288 | 17 | 36 | 43 |
Total number of gains: 54; Total number of losses: 106; Total Number of normals: 328.
Somatic mutations of PARD3:
Generating mutation plots.
Highly correlated genes for PARD3:
Showing top 20/1200 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PARD3 | ZACN | 0.765991 | 3 | 0 | 3 |
| PARD3 | EIF1AY | 0.751064 | 3 | 0 | 3 |
| PARD3 | CLK4 | 0.739315 | 3 | 0 | 3 |
| PARD3 | NHLH2 | 0.732132 | 3 | 0 | 3 |
| PARD3 | KDM5D | 0.731118 | 3 | 0 | 3 |
| PARD3 | EVC2 | 0.730317 | 3 | 0 | 3 |
| PARD3 | ATP6V1H | 0.729085 | 3 | 0 | 3 |
| PARD3 | DDI2 | 0.72873 | 3 | 0 | 3 |
| PARD3 | GPATCH8 | 0.721876 | 3 | 0 | 3 |
| PARD3 | TRIM23 | 0.719608 | 3 | 0 | 3 |
| PARD3 | TYSND1 | 0.714368 | 3 | 0 | 3 |
| PARD3 | TRIM3 | 0.713193 | 3 | 0 | 3 |
| PARD3 | HCFC2 | 0.711888 | 4 | 0 | 3 |
| PARD3 | CASC3 | 0.710417 | 3 | 0 | 3 |
| PARD3 | NAPG | 0.709929 | 3 | 0 | 3 |
| PARD3 | RMND5B | 0.70082 | 11 | 0 | 11 |
| PARD3 | SLC46A2 | 0.700396 | 7 | 0 | 6 |
| PARD3 | JMJD1C | 0.699486 | 5 | 0 | 4 |
| PARD3 | LRRC29 | 0.699313 | 3 | 0 | 3 |
| PARD3 | NAGK | 0.690182 | 11 | 0 | 10 |
For details and further investigation, click here