| Full name: brevican | Alias Symbol: BEHAB|MGC13038|CSPG7 | ||
| Type: protein-coding gene | Cytoband: 1q23.1 | ||
| Entrez ID: 63827 | HGNC ID: HGNC:23059 | Ensembl Gene: ENSG00000132692 | OMIM ID: 600347 |
| Drug and gene relationship at DGIdb | |||
Expression of BCAN:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | BCAN | 63827 | 91920_at | -0.0777 | 0.6964 | |
| GSE20347 | BCAN | 63827 | 91920_at | 0.1207 | 0.2813 | |
| GSE23400 | BCAN | 63827 | 91920_at | -0.1381 | 0.0002 | |
| GSE26886 | BCAN | 63827 | 91920_at | -0.0148 | 0.9148 | |
| GSE29001 | BCAN | 63827 | 91920_at | -0.1316 | 0.4157 | |
| GSE38129 | BCAN | 63827 | 91920_at | -0.0639 | 0.6509 | |
| GSE45670 | BCAN | 63827 | 91920_at | 0.0332 | 0.7195 | |
| GSE53622 | BCAN | 63827 | 7850 | 0.6452 | 0.0008 | |
| GSE53624 | BCAN | 63827 | 7850 | 0.3912 | 0.0031 | |
| GSE63941 | BCAN | 63827 | 91920_at | 0.3146 | 0.0163 | |
| GSE77861 | BCAN | 63827 | 91920_at | -0.1131 | 0.2022 | |
| SRP133303 | BCAN | 63827 | RNAseq | 0.5314 | 0.3591 | |
| TCGA | BCAN | 63827 | RNAseq | 0.2902 | 0.4897 |
Upregulated datasets: 0; Downregulated datasets: 0.
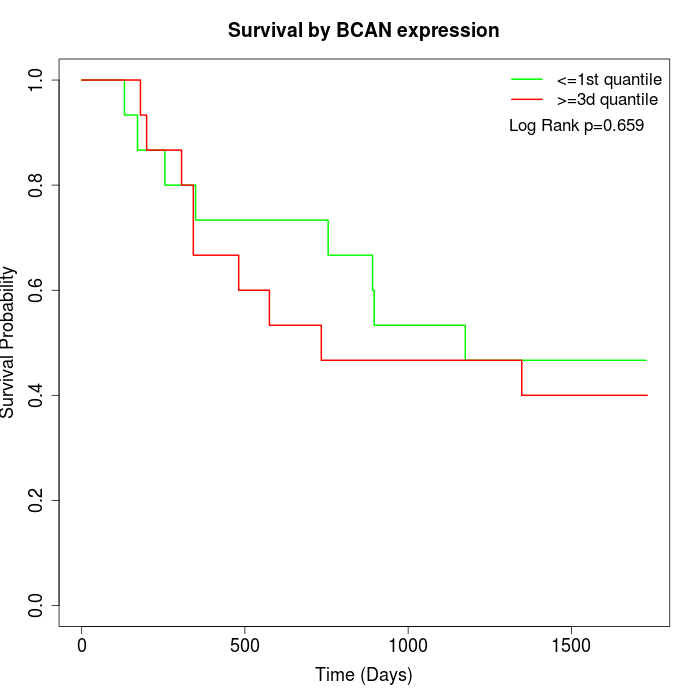
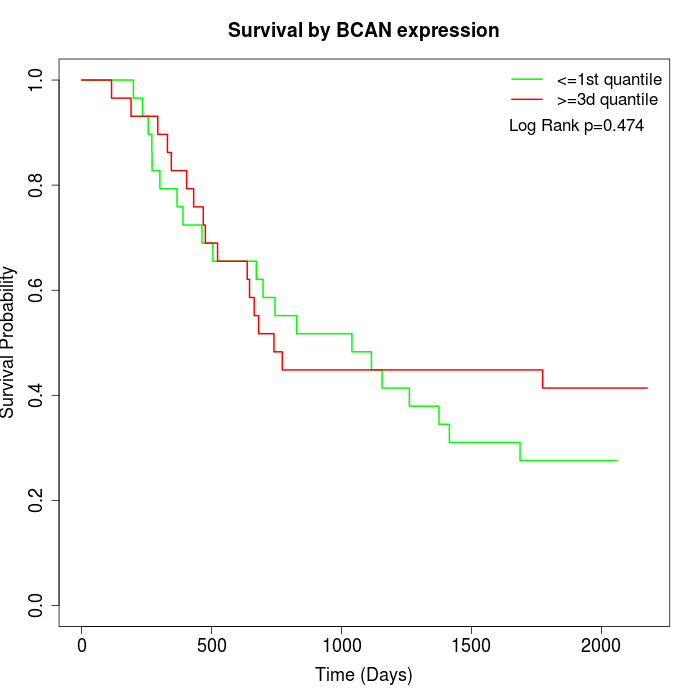
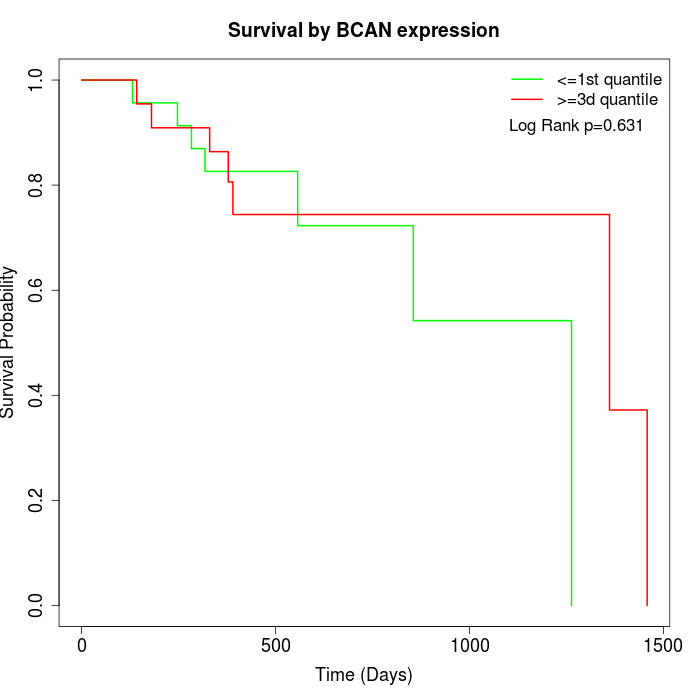
Survival by BCAN expression:
Note: Click image to view full size file.
Copy number change of BCAN:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | BCAN | 63827 | 12 | 0 | 18 | |
| GSE20123 | BCAN | 63827 | 12 | 0 | 18 | |
| GSE43470 | BCAN | 63827 | 7 | 2 | 34 | |
| GSE46452 | BCAN | 63827 | 2 | 1 | 56 | |
| GSE47630 | BCAN | 63827 | 14 | 0 | 26 | |
| GSE54993 | BCAN | 63827 | 0 | 5 | 65 | |
| GSE54994 | BCAN | 63827 | 16 | 0 | 37 | |
| GSE60625 | BCAN | 63827 | 0 | 0 | 11 | |
| GSE74703 | BCAN | 63827 | 7 | 2 | 27 | |
| GSE74704 | BCAN | 63827 | 6 | 0 | 14 | |
| TCGA | BCAN | 63827 | 38 | 2 | 56 |
Total number of gains: 114; Total number of losses: 12; Total Number of normals: 362.
Somatic mutations of BCAN:
Generating mutation plots.
Highly correlated genes for BCAN:
Showing top 20/1453 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| BCAN | CIITA | 0.82981 | 5 | 0 | 5 |
| BCAN | PSG9 | 0.783742 | 3 | 0 | 3 |
| BCAN | NTNG1 | 0.766119 | 4 | 0 | 4 |
| BCAN | OR10C1 | 0.759 | 4 | 0 | 4 |
| BCAN | IL13 | 0.755162 | 5 | 0 | 5 |
| BCAN | KHDRBS2 | 0.751592 | 7 | 0 | 7 |
| BCAN | KCNA1 | 0.748619 | 4 | 0 | 4 |
| BCAN | GPR4 | 0.739742 | 4 | 0 | 4 |
| BCAN | SIGLEC6 | 0.738268 | 6 | 0 | 6 |
| BCAN | FCGR2C | 0.735528 | 3 | 0 | 3 |
| BCAN | CHAT | 0.733595 | 5 | 0 | 5 |
| BCAN | FAM163A | 0.730599 | 4 | 0 | 4 |
| BCAN | ASMT | 0.729502 | 3 | 0 | 3 |
| BCAN | CCL16 | 0.727269 | 4 | 0 | 3 |
| BCAN | RAB3IL1 | 0.727009 | 7 | 0 | 7 |
| BCAN | NXPE4 | 0.726167 | 5 | 0 | 5 |
| BCAN | C8A | 0.725982 | 7 | 0 | 7 |
| BCAN | CHST4 | 0.724352 | 6 | 0 | 6 |
| BCAN | GLP1R | 0.723143 | 7 | 0 | 7 |
| BCAN | MYO1A | 0.722059 | 6 | 0 | 6 |
For details and further investigation, click here