| Full name: potassium voltage-gated channel subfamily A member 1 | Alias Symbol: Kv1.1|RBK1|HUK1|MBK1 | ||
| Type: protein-coding gene | Cytoband: 12p13.32 | ||
| Entrez ID: 3736 | HGNC ID: HGNC:6218 | Ensembl Gene: ENSG00000111262 | OMIM ID: 176260 |
| Drug and gene relationship at DGIdb | |||
Expression of KCNA1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNA1 | 3736 | 208479_at | 0.0539 | 0.8034 | |
| GSE20347 | KCNA1 | 3736 | 208479_at | 0.0640 | 0.4942 | |
| GSE23400 | KCNA1 | 3736 | 208479_at | -0.0925 | 0.0026 | |
| GSE26886 | KCNA1 | 3736 | 208479_at | 0.0062 | 0.9604 | |
| GSE29001 | KCNA1 | 3736 | 208479_at | -0.0089 | 0.9672 | |
| GSE38129 | KCNA1 | 3736 | 208479_at | -0.0306 | 0.7028 | |
| GSE45670 | KCNA1 | 3736 | 208479_at | 0.0773 | 0.3795 | |
| GSE53622 | KCNA1 | 3736 | 29788 | -0.1094 | 0.5477 | |
| GSE53624 | KCNA1 | 3736 | 29788 | 0.2027 | 0.1613 | |
| GSE63941 | KCNA1 | 3736 | 208479_at | 0.0030 | 0.9873 | |
| GSE77861 | KCNA1 | 3736 | 208479_at | -0.0144 | 0.8970 | |
| GSE97050 | KCNA1 | 3736 | A_24_P498652 | 0.0415 | 0.8547 | |
| TCGA | KCNA1 | 3736 | RNAseq | -0.5724 | 0.5473 |
Upregulated datasets: 0; Downregulated datasets: 0.
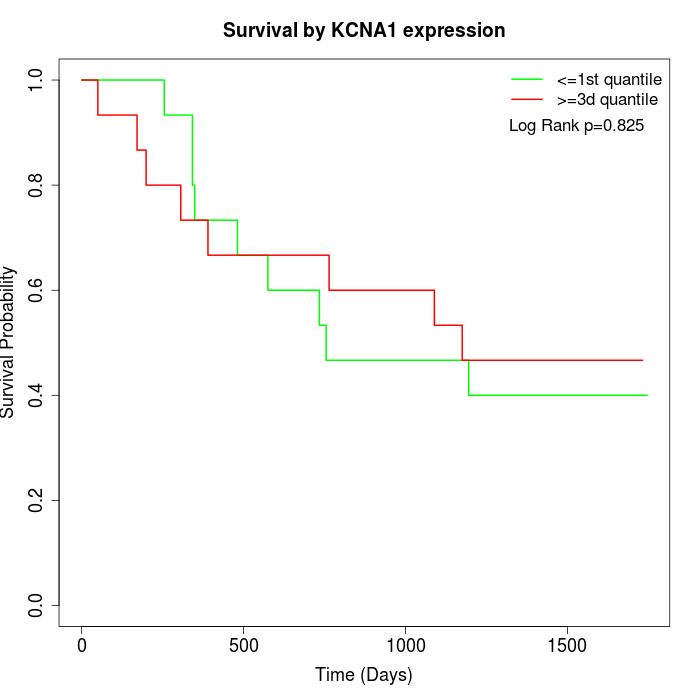
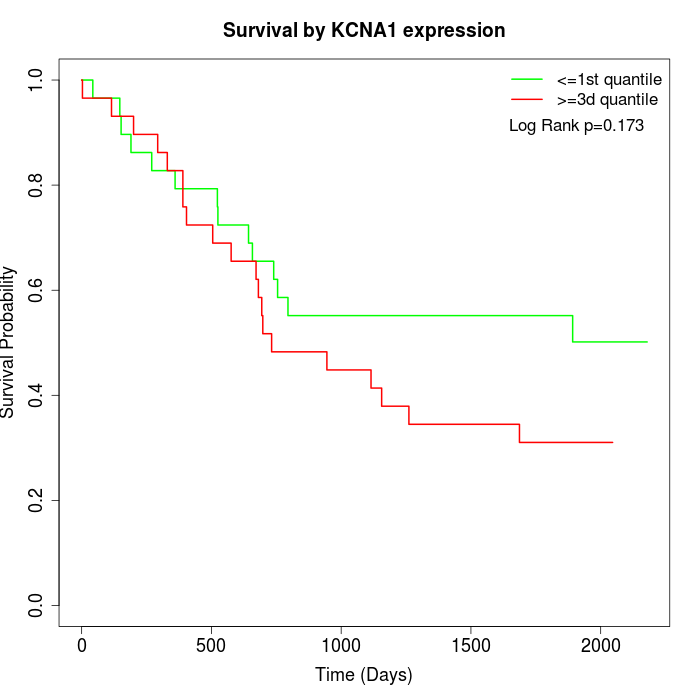
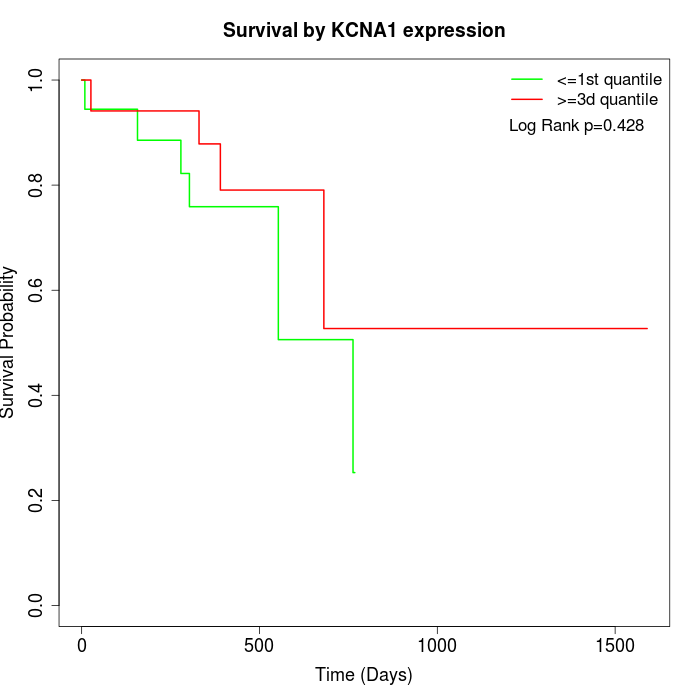
Survival by KCNA1 expression:
Note: Click image to view full size file.
Copy number change of KCNA1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNA1 | 3736 | 6 | 4 | 20 | |
| GSE20123 | KCNA1 | 3736 | 6 | 4 | 20 | |
| GSE43470 | KCNA1 | 3736 | 9 | 3 | 31 | |
| GSE46452 | KCNA1 | 3736 | 10 | 1 | 48 | |
| GSE47630 | KCNA1 | 3736 | 12 | 2 | 26 | |
| GSE54993 | KCNA1 | 3736 | 1 | 10 | 59 | |
| GSE54994 | KCNA1 | 3736 | 10 | 2 | 41 | |
| GSE60625 | KCNA1 | 3736 | 0 | 1 | 10 | |
| GSE74703 | KCNA1 | 3736 | 8 | 2 | 26 | |
| GSE74704 | KCNA1 | 3736 | 4 | 2 | 14 | |
| TCGA | KCNA1 | 3736 | 40 | 6 | 50 |
Total number of gains: 106; Total number of losses: 37; Total Number of normals: 345.
Somatic mutations of KCNA1:
Generating mutation plots.
Highly correlated genes for KCNA1:
Showing top 20/617 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNA1 | BCAN | 0.748619 | 4 | 0 | 4 |
| KCNA1 | GAD2 | 0.742266 | 4 | 0 | 4 |
| KCNA1 | CCR8 | 0.740309 | 3 | 0 | 3 |
| KCNA1 | ZMAT4 | 0.73929 | 3 | 0 | 3 |
| KCNA1 | FCAR | 0.732699 | 4 | 0 | 4 |
| KCNA1 | FCGR2C | 0.726321 | 4 | 0 | 4 |
| KCNA1 | SLC6A12 | 0.724887 | 4 | 0 | 4 |
| KCNA1 | IFNA17 | 0.719564 | 4 | 0 | 3 |
| KCNA1 | GPA33 | 0.717336 | 6 | 0 | 5 |
| KCNA1 | ADAMTS12 | 0.716245 | 5 | 0 | 5 |
| KCNA1 | MEFV | 0.713931 | 4 | 0 | 4 |
| KCNA1 | NTNG1 | 0.699467 | 4 | 0 | 4 |
| KCNA1 | DNMT3L | 0.699465 | 4 | 0 | 4 |
| KCNA1 | RFPL2 | 0.699175 | 5 | 0 | 5 |
| KCNA1 | RNF185-AS1 | 0.693898 | 5 | 0 | 5 |
| KCNA1 | IFT122 | 0.693797 | 4 | 0 | 4 |
| KCNA1 | CDH15 | 0.691914 | 5 | 0 | 5 |
| KCNA1 | SSTR3 | 0.689118 | 4 | 0 | 4 |
| KCNA1 | ART1 | 0.687599 | 4 | 0 | 4 |
| KCNA1 | SYP | 0.687357 | 3 | 0 | 3 |
For details and further investigation, click here