| Full name: butyrophilin like 2 | Alias Symbol: HSBLMHC1|BTL-II|BTN7 | ||
| Type: protein-coding gene | Cytoband: 6p21.32 | ||
| Entrez ID: 56244 | HGNC ID: HGNC:1142 | Ensembl Gene: ENSG00000204290 | OMIM ID: 606000 |
| Drug and gene relationship at DGIdb | |||
Expression of BTNL2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | BTNL2 | 56244 | 221457_s_at | -0.0821 | 0.7718 | |
| GSE20347 | BTNL2 | 56244 | 221457_s_at | -0.0409 | 0.6679 | |
| GSE23400 | BTNL2 | 56244 | 221457_s_at | -0.2848 | 0.0000 | |
| GSE26886 | BTNL2 | 56244 | 221457_s_at | -0.1234 | 0.2602 | |
| GSE29001 | BTNL2 | 56244 | 221457_s_at | -0.2610 | 0.1570 | |
| GSE38129 | BTNL2 | 56244 | 221457_s_at | -0.0985 | 0.3121 | |
| GSE45670 | BTNL2 | 56244 | 221457_s_at | -0.0287 | 0.7981 | |
| GSE53622 | BTNL2 | 56244 | 56231 | 0.2593 | 0.0217 | |
| GSE53624 | BTNL2 | 56244 | 56231 | 0.0696 | 0.4620 | |
| GSE63941 | BTNL2 | 56244 | 221457_s_at | 0.2016 | 0.1767 | |
| GSE77861 | BTNL2 | 56244 | 221457_s_at | -0.2445 | 0.0905 | |
| GSE97050 | BTNL2 | 56244 | A_23_P376686 | -0.0238 | 0.9260 | |
| TCGA | BTNL2 | 56244 | RNAseq | 0.6311 | 0.6719 |
Upregulated datasets: 0; Downregulated datasets: 0.
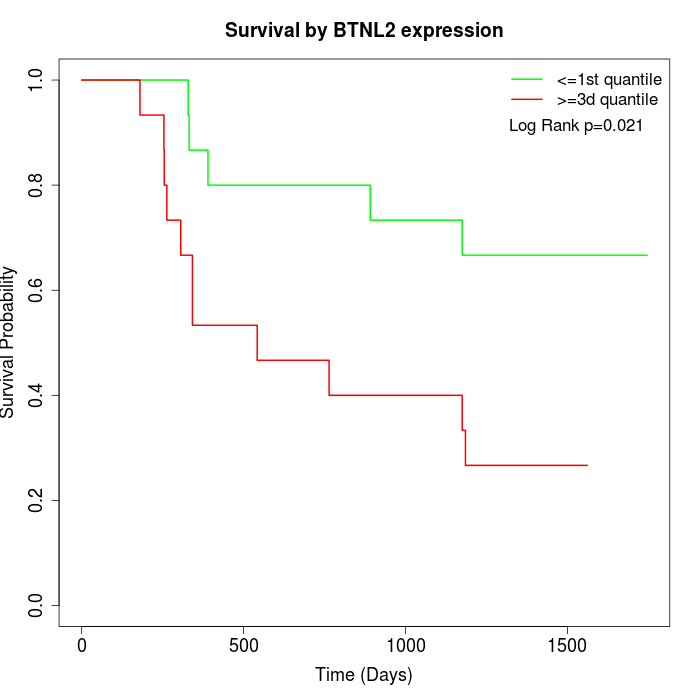
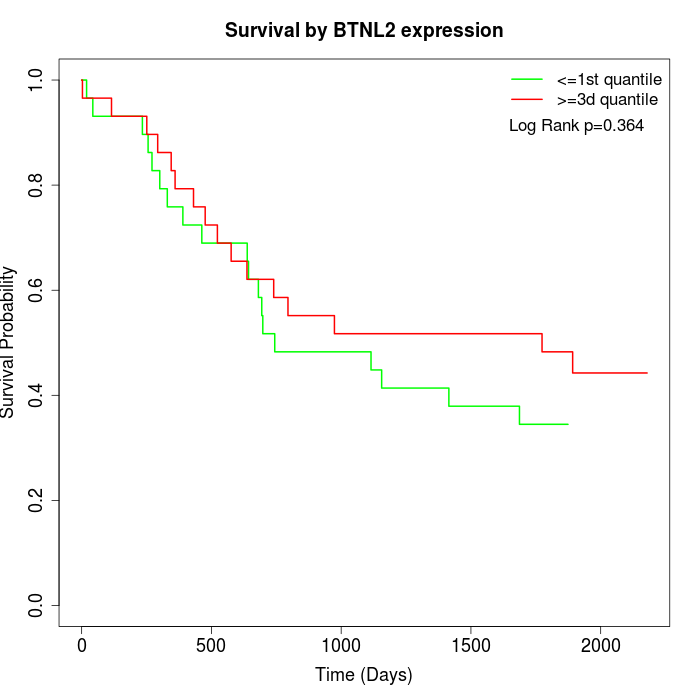
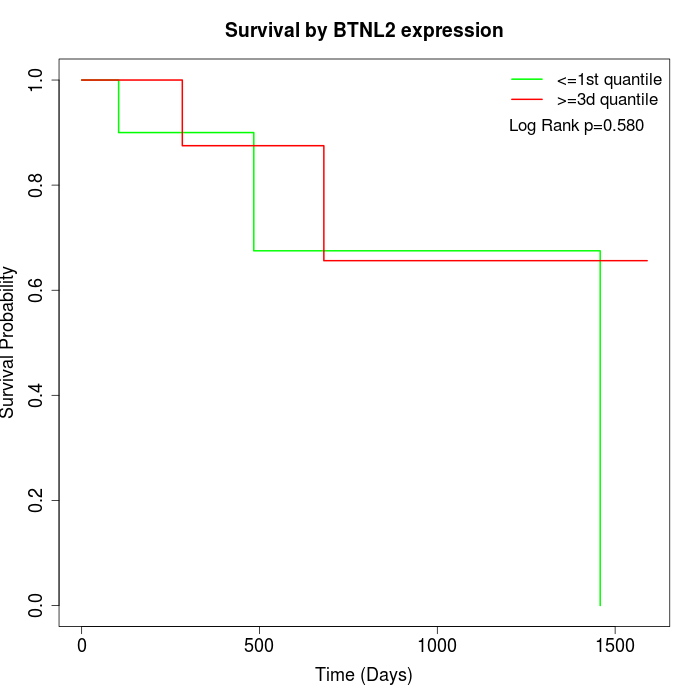
Survival by BTNL2 expression:
Note: Click image to view full size file.
Copy number change of BTNL2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | BTNL2 | 56244 | 6 | 0 | 24 | |
| GSE20123 | BTNL2 | 56244 | 6 | 0 | 24 | |
| GSE43470 | BTNL2 | 56244 | 5 | 1 | 37 | |
| GSE46452 | BTNL2 | 56244 | 1 | 10 | 48 | |
| GSE47630 | BTNL2 | 56244 | 7 | 4 | 29 | |
| GSE54993 | BTNL2 | 56244 | 2 | 1 | 67 | |
| GSE54994 | BTNL2 | 56244 | 11 | 4 | 38 | |
| GSE60625 | BTNL2 | 56244 | 0 | 1 | 10 | |
| GSE74703 | BTNL2 | 56244 | 5 | 0 | 31 | |
| GSE74704 | BTNL2 | 56244 | 2 | 0 | 18 | |
| TCGA | BTNL2 | 56244 | 16 | 16 | 64 |
Total number of gains: 61; Total number of losses: 37; Total Number of normals: 390.
Somatic mutations of BTNL2:
Generating mutation plots.
Highly correlated genes for BTNL2:
Showing top 20/1340 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| BTNL2 | STRC | 0.880498 | 3 | 0 | 3 |
| BTNL2 | SPACA3 | 0.880375 | 3 | 0 | 3 |
| BTNL2 | DCDC1 | 0.870326 | 3 | 0 | 3 |
| BTNL2 | PLA2G2C | 0.849211 | 3 | 0 | 3 |
| BTNL2 | KRTAP10-12 | 0.847583 | 3 | 0 | 3 |
| BTNL2 | TOR2A | 0.826188 | 3 | 0 | 3 |
| BTNL2 | ZNF763 | 0.819512 | 3 | 0 | 3 |
| BTNL2 | OR2T27 | 0.813793 | 3 | 0 | 3 |
| BTNL2 | YY2 | 0.812307 | 3 | 0 | 3 |
| BTNL2 | BBC3 | 0.801258 | 3 | 0 | 3 |
| BTNL2 | C9orf153 | 0.799217 | 3 | 0 | 3 |
| BTNL2 | OR2A5 | 0.794543 | 3 | 0 | 3 |
| BTNL2 | OR4D6 | 0.79358 | 3 | 0 | 3 |
| BTNL2 | ANKDD1A | 0.7918 | 4 | 0 | 4 |
| BTNL2 | FFAR1 | 0.786918 | 3 | 0 | 3 |
| BTNL2 | GCM2 | 0.781771 | 5 | 0 | 4 |
| BTNL2 | OR2T1 | 0.779985 | 3 | 0 | 3 |
| BTNL2 | ASMT | 0.779817 | 3 | 0 | 3 |
| BTNL2 | KRTAP3-1 | 0.779762 | 3 | 0 | 3 |
| BTNL2 | CXorf38 | 0.778911 | 3 | 0 | 3 |
For details and further investigation, click here