| Full name: free fatty acid receptor 1 | Alias Symbol: FFA1R | ||
| Type: protein-coding gene | Cytoband: 19q13.12 | ||
| Entrez ID: 2864 | HGNC ID: HGNC:4498 | Ensembl Gene: ENSG00000126266 | OMIM ID: 603820 |
| Drug and gene relationship at DGIdb | |||
FFAR1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04911 | Insulin secretion |
Expression of FFAR1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | FFAR1 | 2864 | 231761_at | -0.0599 | 0.8451 | |
| GSE26886 | FFAR1 | 2864 | 231761_at | 0.3189 | 0.0195 | |
| GSE45670 | FFAR1 | 2864 | 231761_at | 0.1288 | 0.2424 | |
| GSE53622 | FFAR1 | 2864 | 85985 | 0.2491 | 0.0017 | |
| GSE53624 | FFAR1 | 2864 | 85985 | 0.2888 | 0.0006 | |
| GSE63941 | FFAR1 | 2864 | 231761_at | 0.1298 | 0.4015 | |
| GSE77861 | FFAR1 | 2864 | 231761_at | 0.0211 | 0.8837 | |
| GSE97050 | FFAR1 | 2864 | A_23_P39356 | 0.1270 | 0.6057 |
Upregulated datasets: 0; Downregulated datasets: 0.
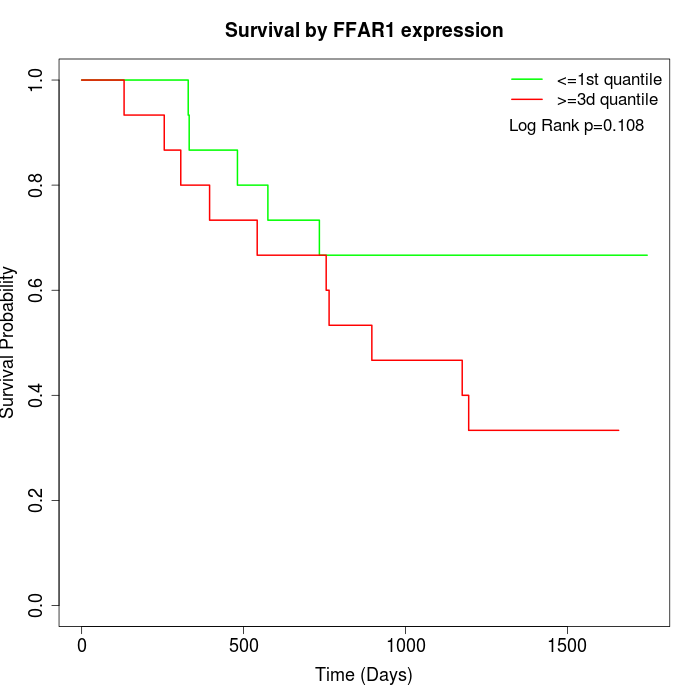
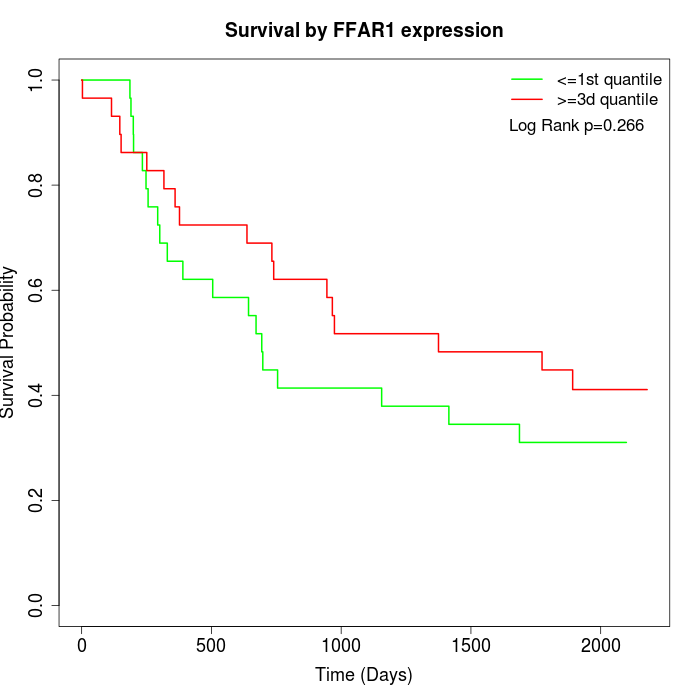
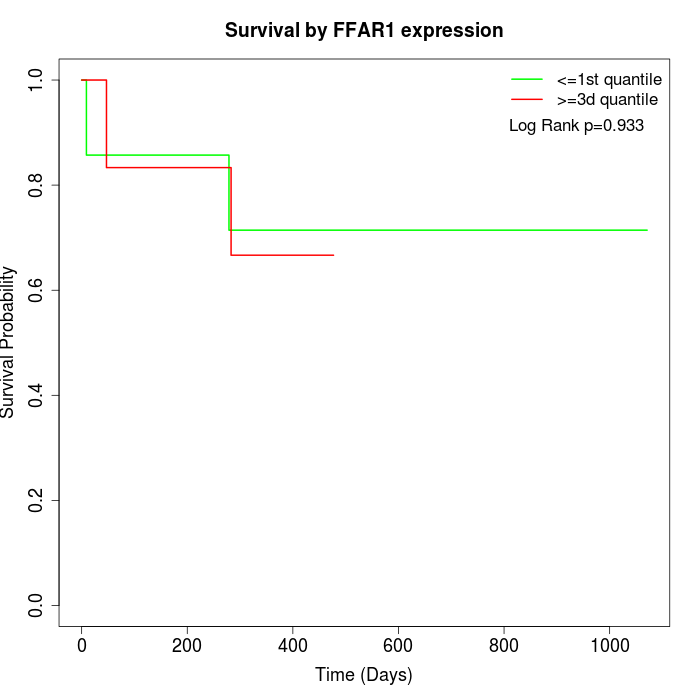
Survival by FFAR1 expression:
Note: Click image to view full size file.
Copy number change of FFAR1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | FFAR1 | 2864 | 6 | 4 | 20 | |
| GSE20123 | FFAR1 | 2864 | 6 | 3 | 21 | |
| GSE43470 | FFAR1 | 2864 | 3 | 7 | 33 | |
| GSE46452 | FFAR1 | 2864 | 48 | 1 | 10 | |
| GSE47630 | FFAR1 | 2864 | 9 | 5 | 26 | |
| GSE54993 | FFAR1 | 2864 | 17 | 3 | 50 | |
| GSE54994 | FFAR1 | 2864 | 8 | 9 | 36 | |
| GSE60625 | FFAR1 | 2864 | 9 | 0 | 2 | |
| GSE74703 | FFAR1 | 2864 | 3 | 4 | 29 | |
| GSE74704 | FFAR1 | 2864 | 6 | 1 | 13 | |
| TCGA | FFAR1 | 2864 | 21 | 11 | 64 |
Total number of gains: 136; Total number of losses: 48; Total Number of normals: 304.
Somatic mutations of FFAR1:
Generating mutation plots.
Highly correlated genes for FFAR1:
Showing top 20/777 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| FFAR1 | YY2 | 0.872217 | 3 | 0 | 3 |
| FFAR1 | C1QL2 | 0.870305 | 3 | 0 | 3 |
| FFAR1 | DCDC1 | 0.867043 | 3 | 0 | 3 |
| FFAR1 | MRGPRG | 0.865782 | 3 | 0 | 3 |
| FFAR1 | PRSS38 | 0.864572 | 3 | 0 | 3 |
| FFAR1 | GRIK4 | 0.8432 | 3 | 0 | 3 |
| FFAR1 | OR2T1 | 0.839744 | 3 | 0 | 3 |
| FFAR1 | OR2B11 | 0.828882 | 3 | 0 | 3 |
| FFAR1 | DEFB121 | 0.826937 | 4 | 0 | 4 |
| FFAR1 | CHST10 | 0.821789 | 3 | 0 | 3 |
| FFAR1 | PABPN1L | 0.821453 | 3 | 0 | 3 |
| FFAR1 | ABTB1 | 0.818449 | 3 | 0 | 3 |
| FFAR1 | PLA2G2C | 0.817866 | 3 | 0 | 3 |
| FFAR1 | C19orf71 | 0.81406 | 3 | 0 | 3 |
| FFAR1 | C9orf153 | 0.813686 | 3 | 0 | 3 |
| FFAR1 | STRC | 0.813487 | 3 | 0 | 3 |
| FFAR1 | BBC3 | 0.812069 | 3 | 0 | 3 |
| FFAR1 | LHB | 0.808316 | 3 | 0 | 3 |
| FFAR1 | OR2A5 | 0.8072 | 3 | 0 | 3 |
| FFAR1 | OTOG | 0.802186 | 3 | 0 | 3 |
For details and further investigation, click here