| Full name: complement C8 gamma chain | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 9q34.3 | ||
| Entrez ID: 733 | HGNC ID: HGNC:1354 | Ensembl Gene: ENSG00000176919 | OMIM ID: 120930 |
| Drug and gene relationship at DGIdb | |||
Expression of C8G:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | C8G | 733 | 210324_at | 0.1921 | 0.4451 | |
| GSE20347 | C8G | 733 | 210324_at | -0.0210 | 0.7771 | |
| GSE23400 | C8G | 733 | 210324_at | -0.0296 | 0.4737 | |
| GSE26886 | C8G | 733 | 210324_at | 0.2098 | 0.0651 | |
| GSE29001 | C8G | 733 | 210324_at | 0.0568 | 0.6487 | |
| GSE38129 | C8G | 733 | 210324_at | 0.0594 | 0.3713 | |
| GSE45670 | C8G | 733 | 210324_at | 0.1206 | 0.2344 | |
| GSE53622 | C8G | 733 | 73178 | -0.3273 | 0.0000 | |
| GSE53624 | C8G | 733 | 73178 | -0.4770 | 0.0000 | |
| GSE63941 | C8G | 733 | 210324_at | 0.5345 | 0.0054 | |
| GSE77861 | C8G | 733 | 210324_at | -0.0434 | 0.7954 | |
| GSE97050 | C8G | 733 | A_33_P3319006 | -0.1072 | 0.6733 | |
| SRP219564 | C8G | 733 | RNAseq | 0.2555 | 0.6844 | |
| TCGA | C8G | 733 | RNAseq | 0.1089 | 0.8240 |
Upregulated datasets: 0; Downregulated datasets: 0.
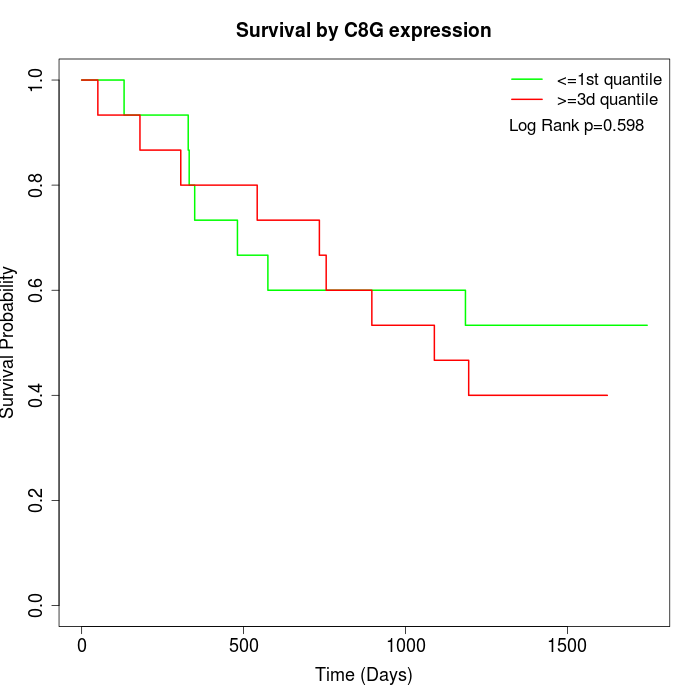
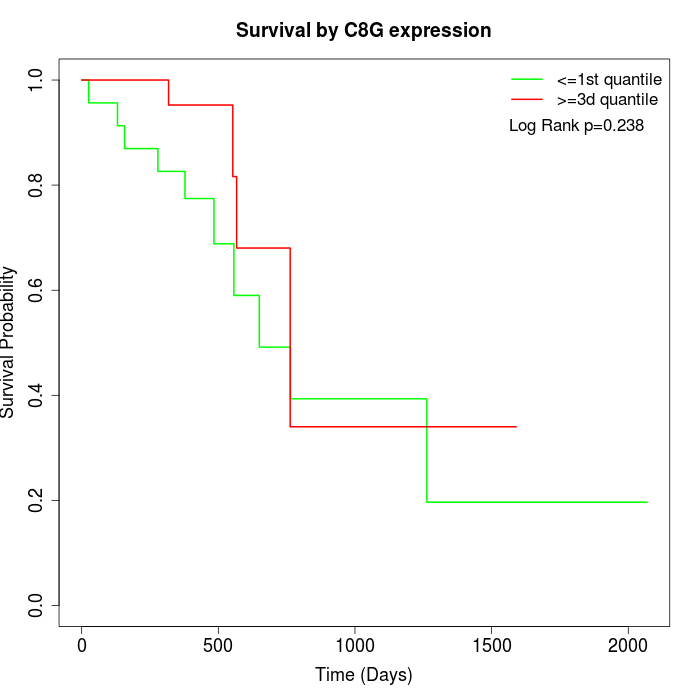
Survival by C8G expression:
Note: Click image to view full size file.
Copy number change of C8G:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | C8G | 733 | 5 | 7 | 18 | |
| GSE20123 | C8G | 733 | 5 | 7 | 18 | |
| GSE43470 | C8G | 733 | 3 | 7 | 33 | |
| GSE46452 | C8G | 733 | 6 | 13 | 40 | |
| GSE47630 | C8G | 733 | 6 | 15 | 19 | |
| GSE54993 | C8G | 733 | 3 | 3 | 64 | |
| GSE54994 | C8G | 733 | 12 | 8 | 33 | |
| GSE60625 | C8G | 733 | 0 | 0 | 11 | |
| GSE74703 | C8G | 733 | 3 | 5 | 28 | |
| GSE74704 | C8G | 733 | 3 | 5 | 12 | |
| TCGA | C8G | 733 | 29 | 23 | 44 |
Total number of gains: 75; Total number of losses: 93; Total Number of normals: 320.
Somatic mutations of C8G:
Generating mutation plots.
Highly correlated genes for C8G:
Showing top 20/445 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| C8G | GDF7 | 0.817408 | 3 | 0 | 3 |
| C8G | MYLK3 | 0.788989 | 3 | 0 | 3 |
| C8G | KRTAP6-1 | 0.78503 | 3 | 0 | 3 |
| C8G | SLC34A1 | 0.782758 | 3 | 0 | 3 |
| C8G | CLEC18C | 0.781564 | 3 | 0 | 3 |
| C8G | ANKRD33B | 0.768198 | 3 | 0 | 3 |
| C8G | DCD | 0.767253 | 3 | 0 | 3 |
| C8G | BARHL2 | 0.766378 | 3 | 0 | 3 |
| C8G | MFSD2B | 0.765576 | 3 | 0 | 3 |
| C8G | CHD5 | 0.760047 | 4 | 0 | 4 |
| C8G | FOXL2 | 0.759304 | 3 | 0 | 3 |
| C8G | C20orf141 | 0.755378 | 4 | 0 | 4 |
| C8G | KBTBD13 | 0.755076 | 3 | 0 | 3 |
| C8G | C9orf106 | 0.751744 | 3 | 0 | 3 |
| C8G | TPRX1 | 0.750538 | 3 | 0 | 3 |
| C8G | KRTAP20-1 | 0.750151 | 3 | 0 | 3 |
| C8G | CLRN2 | 0.745623 | 3 | 0 | 3 |
| C8G | ZNF497 | 0.743364 | 3 | 0 | 3 |
| C8G | YY2 | 0.738191 | 3 | 0 | 3 |
| C8G | KRT77 | 0.736977 | 3 | 0 | 3 |
For details and further investigation, click here