| Full name: carbonic anhydrase 6 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 1p36.23 | ||
| Entrez ID: 765 | HGNC ID: HGNC:1380 | Ensembl Gene: ENSG00000131686 | OMIM ID: 114780 |
| Drug and gene relationship at DGIdb | |||
Expression of CA6:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CA6 | 765 | 206873_at | 0.1293 | 0.4284 | |
| GSE20347 | CA6 | 765 | 206873_at | 0.2184 | 0.2147 | |
| GSE23400 | CA6 | 765 | 206873_at | -0.0283 | 0.4651 | |
| GSE26886 | CA6 | 765 | 206873_at | 0.1114 | 0.3534 | |
| GSE29001 | CA6 | 765 | 206873_at | 0.2309 | 0.6095 | |
| GSE38129 | CA6 | 765 | 206873_at | 0.2724 | 0.2339 | |
| GSE45670 | CA6 | 765 | 206873_at | 0.2250 | 0.0986 | |
| GSE53622 | CA6 | 765 | 40137 | 0.4983 | 0.0000 | |
| GSE53624 | CA6 | 765 | 40137 | 0.1908 | 0.0428 | |
| GSE63941 | CA6 | 765 | 206873_at | 0.2272 | 0.4208 | |
| GSE77861 | CA6 | 765 | 206873_at | 0.0121 | 0.9243 | |
| GSE97050 | CA6 | 765 | A_33_P3334398 | 0.1689 | 0.4352 | |
| TCGA | CA6 | 765 | RNAseq | 2.4452 | 0.0843 |
Upregulated datasets: 0; Downregulated datasets: 0.
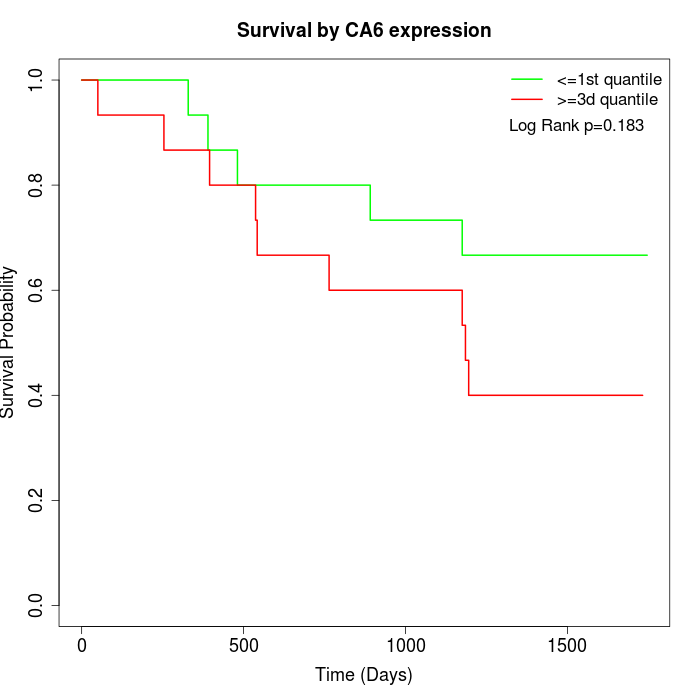
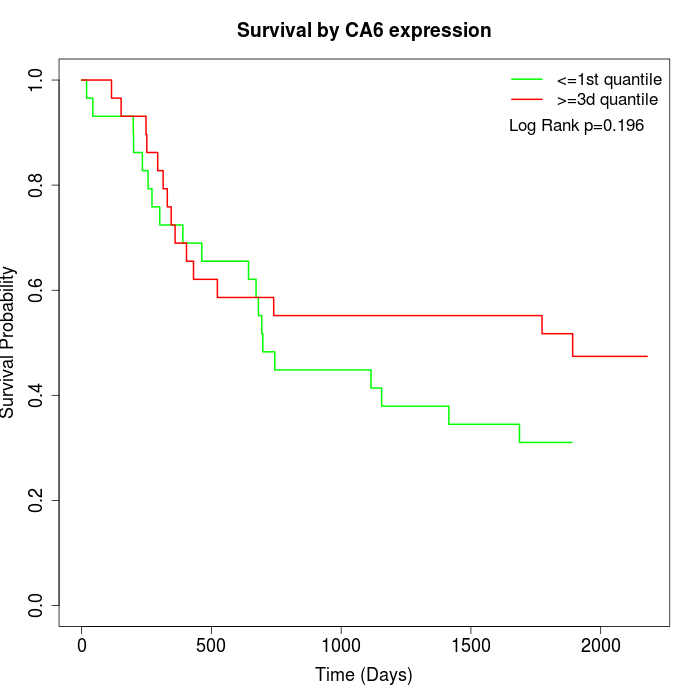
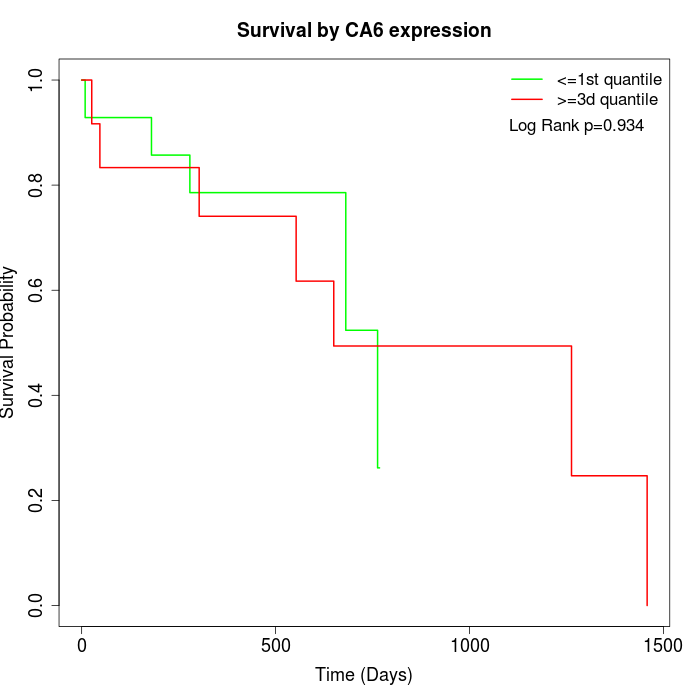
Survival by CA6 expression:
Note: Click image to view full size file.
Copy number change of CA6:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CA6 | 765 | 1 | 5 | 24 | |
| GSE20123 | CA6 | 765 | 1 | 3 | 26 | |
| GSE43470 | CA6 | 765 | 6 | 5 | 32 | |
| GSE46452 | CA6 | 765 | 7 | 1 | 51 | |
| GSE47630 | CA6 | 765 | 8 | 4 | 28 | |
| GSE54993 | CA6 | 765 | 3 | 2 | 65 | |
| GSE54994 | CA6 | 765 | 12 | 4 | 37 | |
| GSE60625 | CA6 | 765 | 0 | 0 | 11 | |
| GSE74703 | CA6 | 765 | 5 | 3 | 28 | |
| GSE74704 | CA6 | 765 | 1 | 0 | 19 | |
| TCGA | CA6 | 765 | 11 | 24 | 61 |
Total number of gains: 55; Total number of losses: 51; Total Number of normals: 382.
Somatic mutations of CA6:
Generating mutation plots.
Highly correlated genes for CA6:
Showing top 20/400 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CA6 | FBRSL1 | 0.828895 | 3 | 0 | 3 |
| CA6 | STRC | 0.815157 | 3 | 0 | 3 |
| CA6 | DCDC1 | 0.814563 | 3 | 0 | 3 |
| CA6 | ANXA10 | 0.805763 | 3 | 0 | 3 |
| CA6 | CLTCL1 | 0.805035 | 3 | 0 | 3 |
| CA6 | ZNF527 | 0.789027 | 3 | 0 | 3 |
| CA6 | CIB3 | 0.787767 | 3 | 0 | 3 |
| CA6 | WDR87 | 0.779461 | 3 | 0 | 3 |
| CA6 | C2CD4D | 0.777955 | 3 | 0 | 3 |
| CA6 | TP73 | 0.77645 | 4 | 0 | 4 |
| CA6 | ERC2 | 0.774781 | 3 | 0 | 3 |
| CA6 | PLA2G2C | 0.769658 | 3 | 0 | 3 |
| CA6 | C20orf85 | 0.76465 | 3 | 0 | 3 |
| CA6 | RASL10B | 0.761725 | 3 | 0 | 3 |
| CA6 | TOR2A | 0.761572 | 3 | 0 | 3 |
| CA6 | CELF5 | 0.761543 | 3 | 0 | 3 |
| CA6 | BTNL2 | 0.751653 | 4 | 0 | 4 |
| CA6 | MRGPRG | 0.743849 | 3 | 0 | 3 |
| CA6 | YY2 | 0.742115 | 3 | 0 | 3 |
| CA6 | OR2C3 | 0.73934 | 3 | 0 | 3 |
For details and further investigation, click here