| Full name: CUGBP Elav-like family member 5 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 19p13.3 | ||
| Entrez ID: 60680 | HGNC ID: HGNC:14058 | Ensembl Gene: ENSG00000161082 | OMIM ID: 612680 |
| Drug and gene relationship at DGIdb | |||
Expression of CELF5:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CELF5 | 60680 | 231744_at | -0.1080 | 0.6767 | |
| GSE26886 | CELF5 | 60680 | 230497_at | 0.1465 | 0.5236 | |
| GSE45670 | CELF5 | 60680 | 230497_at | 0.0572 | 0.6175 | |
| GSE53622 | CELF5 | 60680 | 105687 | 0.0301 | 0.7471 | |
| GSE53624 | CELF5 | 60680 | 105687 | -0.2735 | 0.0111 | |
| GSE63941 | CELF5 | 60680 | 231744_at | 0.1863 | 0.1894 | |
| GSE77861 | CELF5 | 60680 | 230497_at | -0.0954 | 0.3074 | |
| GSE97050 | CELF5 | 60680 | A_33_P3386156 | 0.0375 | 0.9029 | |
| SRP133303 | CELF5 | 60680 | RNAseq | 0.1219 | 0.4696 | |
| SRP219564 | CELF5 | 60680 | RNAseq | 0.6323 | 0.2549 | |
| TCGA | CELF5 | 60680 | RNAseq | -0.9817 | 0.0710 |
Upregulated datasets: 0; Downregulated datasets: 0.
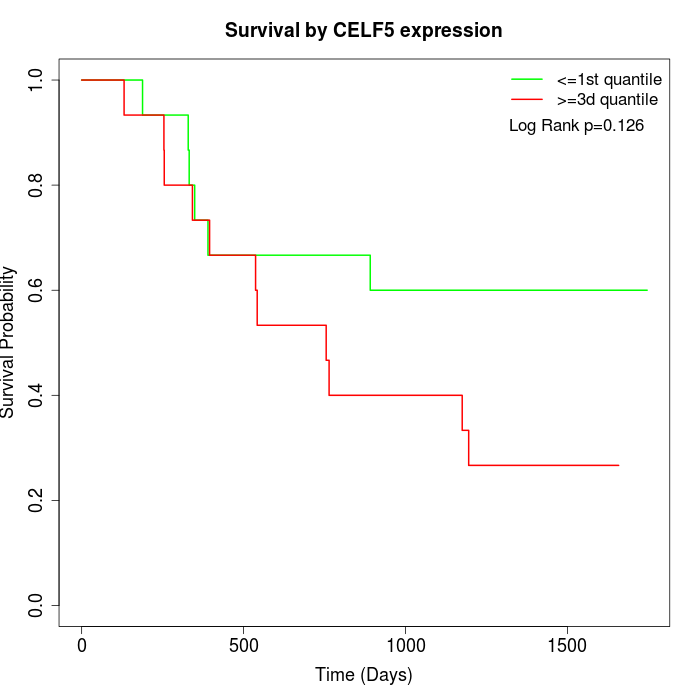
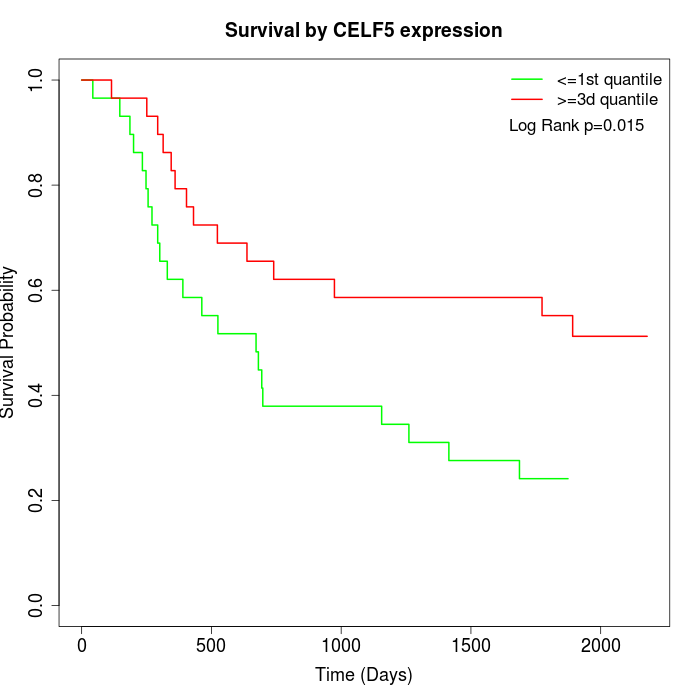
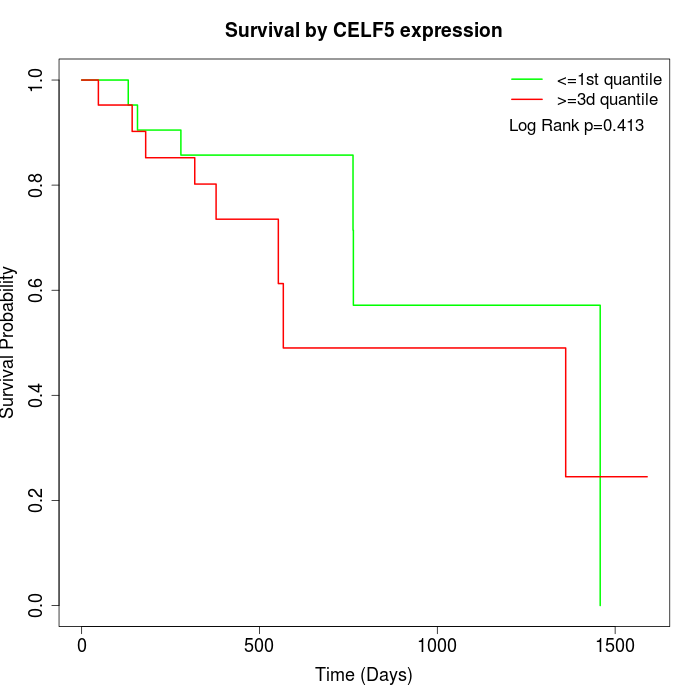
Survival by CELF5 expression:
Note: Click image to view full size file.
Copy number change of CELF5:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CELF5 | 60680 | 5 | 4 | 21 | |
| GSE20123 | CELF5 | 60680 | 3 | 4 | 23 | |
| GSE43470 | CELF5 | 60680 | 1 | 9 | 33 | |
| GSE46452 | CELF5 | 60680 | 47 | 1 | 11 | |
| GSE47630 | CELF5 | 60680 | 5 | 7 | 28 | |
| GSE54993 | CELF5 | 60680 | 16 | 3 | 51 | |
| GSE54994 | CELF5 | 60680 | 8 | 16 | 29 | |
| GSE60625 | CELF5 | 60680 | 9 | 0 | 2 | |
| GSE74703 | CELF5 | 60680 | 1 | 6 | 29 | |
| GSE74704 | CELF5 | 60680 | 1 | 3 | 16 | |
| TCGA | CELF5 | 60680 | 10 | 20 | 66 |
Total number of gains: 106; Total number of losses: 73; Total Number of normals: 309.
Somatic mutations of CELF5:
Generating mutation plots.
Highly correlated genes for CELF5:
Showing top 20/354 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CELF5 | KCP | 0.873755 | 3 | 0 | 3 |
| CELF5 | STRC | 0.860413 | 3 | 0 | 3 |
| CELF5 | GRIK4 | 0.850124 | 3 | 0 | 3 |
| CELF5 | KIRREL2 | 0.84631 | 3 | 0 | 3 |
| CELF5 | FZD9 | 0.835224 | 3 | 0 | 3 |
| CELF5 | YY2 | 0.829867 | 3 | 0 | 3 |
| CELF5 | GFRA4 | 0.82626 | 3 | 0 | 3 |
| CELF5 | PLA2G2C | 0.826124 | 3 | 0 | 3 |
| CELF5 | PTAFR | 0.821878 | 3 | 0 | 3 |
| CELF5 | NOBOX | 0.817283 | 3 | 0 | 3 |
| CELF5 | KRTAP4-8 | 0.816114 | 3 | 0 | 3 |
| CELF5 | TCTE1 | 0.811075 | 3 | 0 | 3 |
| CELF5 | TOR2A | 0.810987 | 3 | 0 | 3 |
| CELF5 | SLC26A1 | 0.806092 | 3 | 0 | 3 |
| CELF5 | DCDC1 | 0.804972 | 3 | 0 | 3 |
| CELF5 | OR10AD1 | 0.802558 | 3 | 0 | 3 |
| CELF5 | GPR152 | 0.799288 | 3 | 0 | 3 |
| CELF5 | CXorf38 | 0.79907 | 3 | 0 | 3 |
| CELF5 | TUBA3D | 0.797807 | 3 | 0 | 3 |
| CELF5 | MRGPRE | 0.795825 | 3 | 0 | 3 |
For details and further investigation, click here