| Full name: calcium voltage-gated channel auxiliary subunit alpha2delta 1 | Alias Symbol: lncRNA-N3 | ||
| Type: protein-coding gene | Cytoband: 7q21.11 | ||
| Entrez ID: 781 | HGNC ID: HGNC:1399 | Ensembl Gene: ENSG00000153956 | OMIM ID: 114204 |
| Drug and gene relationship at DGIdb | |||
CACNA2D1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04010 | MAPK signaling pathway | |
| hsa04261 | Adrenergic signaling in cardiomyocytes | |
| hsa04921 | Oxytocin signaling pathway |
Expression of CACNA2D1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CACNA2D1 | 781 | 227623_at | -0.7420 | 0.6143 | |
| GSE20347 | CACNA2D1 | 781 | 207050_at | 0.0653 | 0.2706 | |
| GSE23400 | CACNA2D1 | 781 | 207050_at | -0.0065 | 0.8497 | |
| GSE26886 | CACNA2D1 | 781 | 207050_at | -0.1097 | 0.3086 | |
| GSE29001 | CACNA2D1 | 781 | 207050_at | -0.0969 | 0.4706 | |
| GSE38129 | CACNA2D1 | 781 | 207050_at | -0.0119 | 0.8800 | |
| GSE45670 | CACNA2D1 | 781 | 227623_at | -0.6409 | 0.2827 | |
| GSE53622 | CACNA2D1 | 781 | 12761 | -0.2298 | 0.2192 | |
| GSE53624 | CACNA2D1 | 781 | 12761 | 0.1936 | 0.1433 | |
| GSE63941 | CACNA2D1 | 781 | 227623_at | -4.5596 | 0.0145 | |
| GSE77861 | CACNA2D1 | 781 | 207050_at | -0.0833 | 0.2862 | |
| GSE97050 | CACNA2D1 | 781 | A_33_P3232478 | -0.6242 | 0.1292 | |
| SRP007169 | CACNA2D1 | 781 | RNAseq | 2.9430 | 0.0000 | |
| SRP008496 | CACNA2D1 | 781 | RNAseq | 2.7858 | 0.0000 | |
| SRP064894 | CACNA2D1 | 781 | RNAseq | 0.4317 | 0.1762 | |
| SRP133303 | CACNA2D1 | 781 | RNAseq | 0.6684 | 0.0972 | |
| SRP159526 | CACNA2D1 | 781 | RNAseq | 0.3663 | 0.2625 | |
| SRP193095 | CACNA2D1 | 781 | RNAseq | 0.7690 | 0.0049 | |
| SRP219564 | CACNA2D1 | 781 | RNAseq | -0.3647 | 0.6518 | |
| TCGA | CACNA2D1 | 781 | RNAseq | -0.4005 | 0.2249 |
Upregulated datasets: 2; Downregulated datasets: 1.
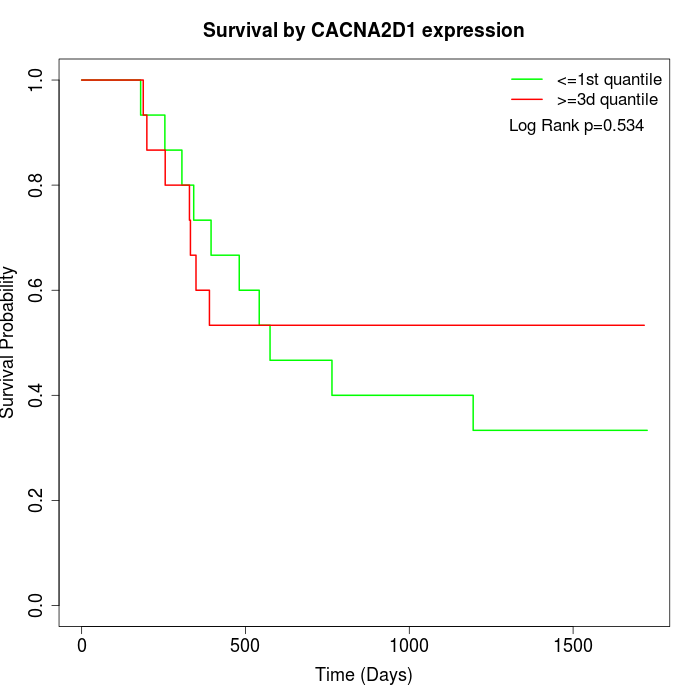
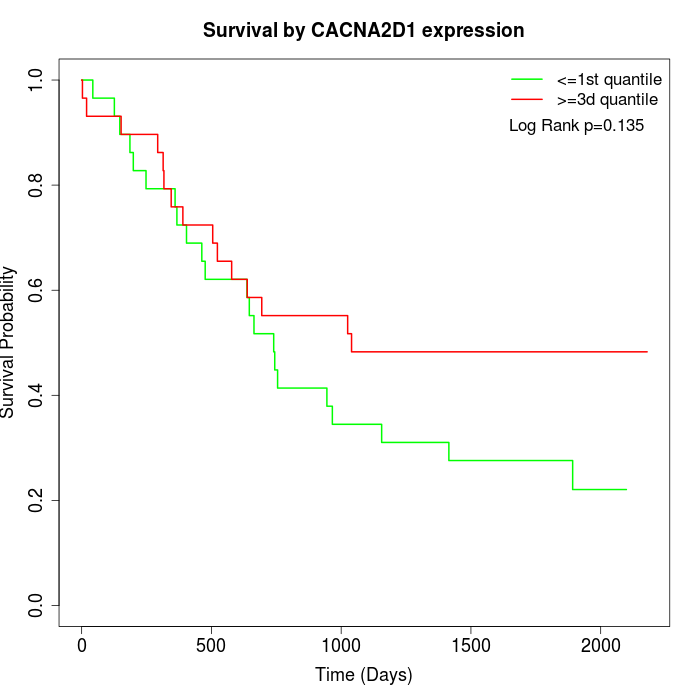
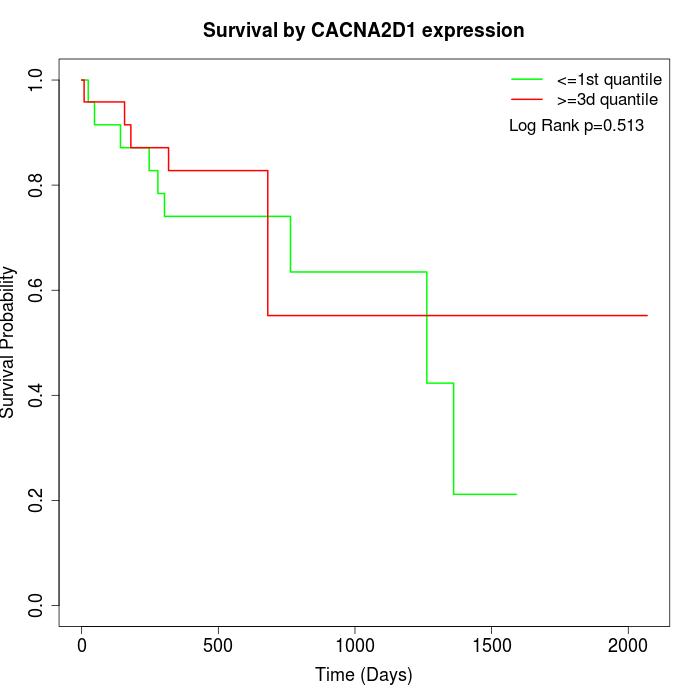
Survival by CACNA2D1 expression:
Note: Click image to view full size file.
Copy number change of CACNA2D1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CACNA2D1 | 781 | 12 | 1 | 17 | |
| GSE20123 | CACNA2D1 | 781 | 11 | 1 | 18 | |
| GSE43470 | CACNA2D1 | 781 | 4 | 1 | 38 | |
| GSE46452 | CACNA2D1 | 781 | 11 | 0 | 48 | |
| GSE47630 | CACNA2D1 | 781 | 7 | 3 | 30 | |
| GSE54993 | CACNA2D1 | 781 | 1 | 7 | 62 | |
| GSE54994 | CACNA2D1 | 781 | 16 | 3 | 34 | |
| GSE60625 | CACNA2D1 | 781 | 0 | 0 | 11 | |
| GSE74703 | CACNA2D1 | 781 | 3 | 1 | 32 | |
| GSE74704 | CACNA2D1 | 781 | 7 | 1 | 12 | |
| TCGA | CACNA2D1 | 781 | 47 | 5 | 44 |
Total number of gains: 119; Total number of losses: 23; Total Number of normals: 346.
Somatic mutations of CACNA2D1:
Generating mutation plots.
Highly correlated genes for CACNA2D1:
Showing top 20/427 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CACNA2D1 | CXCL12 | 0.738301 | 3 | 0 | 3 |
| CACNA2D1 | ZNF583 | 0.723145 | 3 | 0 | 3 |
| CACNA2D1 | ZNF610 | 0.721788 | 3 | 0 | 3 |
| CACNA2D1 | ZNF382 | 0.718333 | 4 | 0 | 4 |
| CACNA2D1 | MICA | 0.715678 | 3 | 0 | 3 |
| CACNA2D1 | PLAC9 | 0.711351 | 3 | 0 | 3 |
| CACNA2D1 | FNDC5 | 0.707898 | 3 | 0 | 3 |
| CACNA2D1 | KLHDC8B | 0.704022 | 3 | 0 | 3 |
| CACNA2D1 | ZNF132 | 0.698347 | 4 | 0 | 4 |
| CACNA2D1 | MAOA | 0.697903 | 3 | 0 | 3 |
| CACNA2D1 | KBTBD8 | 0.69655 | 3 | 0 | 3 |
| CACNA2D1 | CCDC87 | 0.693008 | 3 | 0 | 3 |
| CACNA2D1 | PER3 | 0.690581 | 3 | 0 | 3 |
| CACNA2D1 | IFFO1 | 0.69029 | 4 | 0 | 4 |
| CACNA2D1 | BVES | 0.688765 | 4 | 0 | 4 |
| CACNA2D1 | IL1RAPL2 | 0.687006 | 3 | 0 | 3 |
| CACNA2D1 | GPC6 | 0.686143 | 3 | 0 | 3 |
| CACNA2D1 | CACNB1 | 0.686013 | 4 | 0 | 4 |
| CACNA2D1 | KLF2 | 0.685386 | 4 | 0 | 3 |
| CACNA2D1 | ANGPTL6 | 0.685327 | 3 | 0 | 3 |
For details and further investigation, click here