| Full name: MHC class I polypeptide-related sequence A | Alias Symbol: PERB11.1 | ||
| Type: protein-coding gene | Cytoband: 6p21.33 | ||
| Entrez ID: 100507436 | HGNC ID: HGNC:7090 | Ensembl Gene: ENSG00000204520 | OMIM ID: 600169 |
| Drug and gene relationship at DGIdb | |||
MICA involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04650 | Natural killer cell mediated cytotoxicity |
Expression of MICA:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MICA | 100507436 | 205904_at | 0.0324 | 0.9779 | |
| GSE20347 | MICA | 100507436 | 205904_at | 0.4200 | 0.0396 | |
| GSE23400 | MICA | 100507436 | 205904_at | 0.1115 | 0.1757 | |
| GSE26886 | MICA | 100507436 | 205904_at | 0.9383 | 0.0005 | |
| GSE29001 | MICA | 100507436 | 205904_at | 0.2556 | 0.5905 | |
| GSE38129 | MICA | 100507436 | 205904_at | 0.3450 | 0.0679 | |
| GSE45670 | MICA | 100507436 | 205904_at | 0.0565 | 0.8826 | |
| GSE53622 | MICA | 100507436 | 56453 | 0.3877 | 0.0035 | |
| GSE53624 | MICA | 100507436 | 56453 | 0.4361 | 0.0000 | |
| GSE63941 | MICA | 100507436 | 205904_at | -1.7536 | 0.0069 | |
| GSE77861 | MICA | 100507436 | 205904_at | 0.2292 | 0.5524 | |
| GSE97050 | MICA | 100507436 | A_33_P3368453 | -0.2600 | 0.2727 | |
| SRP007169 | MICA | 100507436 | RNAseq | 0.5992 | 0.1920 | |
| SRP008496 | MICA | 100507436 | RNAseq | 0.4297 | 0.1789 | |
| SRP064894 | MICA | 100507436 | RNAseq | 0.2679 | 0.2035 | |
| SRP133303 | MICA | 100507436 | RNAseq | 0.0696 | 0.8311 | |
| SRP159526 | MICA | 100507436 | RNAseq | 0.3220 | 0.3637 | |
| SRP193095 | MICA | 100507436 | RNAseq | 0.4217 | 0.0031 | |
| SRP219564 | MICA | 100507436 | RNAseq | -0.2511 | 0.5840 | |
| TCGA | MICA | 100507436 | RNAseq | -0.1332 | 0.0452 |
Upregulated datasets: 0; Downregulated datasets: 1.
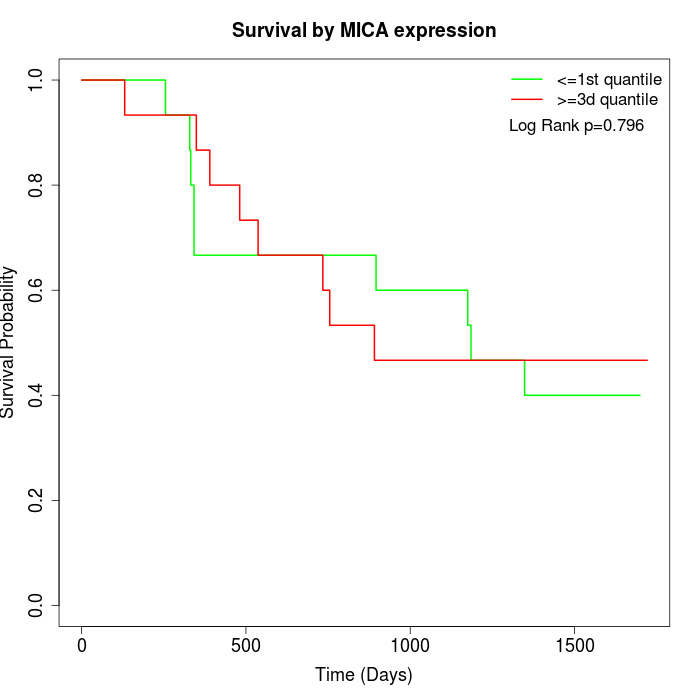
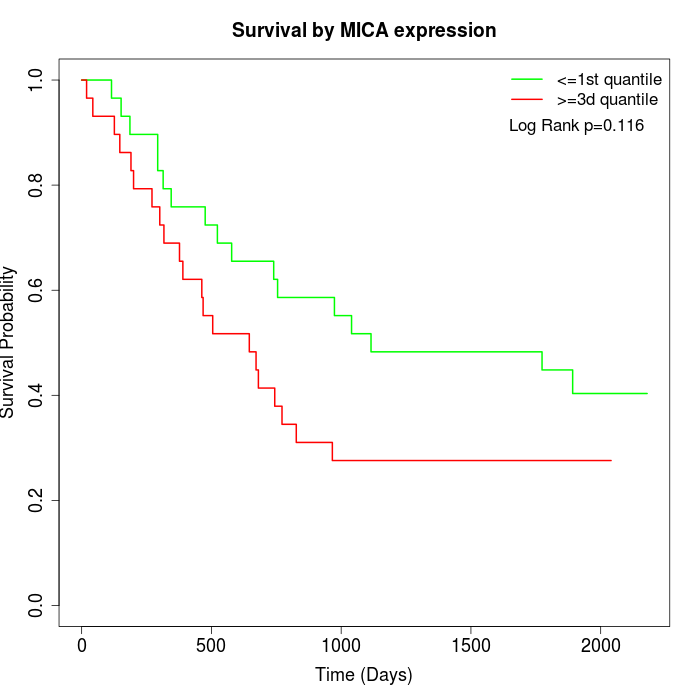
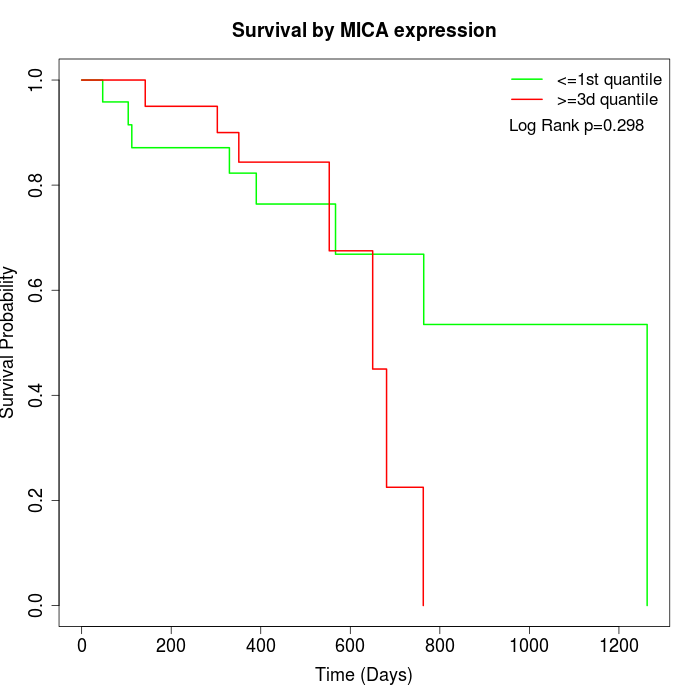
Survival by MICA expression:
Note: Click image to view full size file.
Copy number change of MICA:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MICA | 100507436 | 5 | 1 | 24 | |
| GSE20123 | MICA | 100507436 | 5 | 1 | 24 | |
| GSE43470 | MICA | 100507436 | 5 | 1 | 37 | |
| GSE46452 | MICA | 100507436 | 1 | 10 | 48 | |
| GSE47630 | MICA | 100507436 | 7 | 3 | 30 | |
| GSE54993 | MICA | 100507436 | 2 | 1 | 67 | |
| GSE54994 | MICA | 100507436 | 11 | 4 | 38 | |
| GSE60625 | MICA | 100507436 | 0 | 1 | 10 | |
| GSE74703 | MICA | 100507436 | 5 | 0 | 31 | |
| GSE74704 | MICA | 100507436 | 2 | 0 | 18 | |
| TCGA | MICA | 100507436 | 16 | 16 | 64 |
Total number of gains: 59; Total number of losses: 38; Total Number of normals: 391.
Somatic mutations of MICA:
Generating mutation plots.
Highly correlated genes for MICA:
Showing top 20/388 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MICA | SAR1A | 0.745399 | 3 | 0 | 3 |
| MICA | ZNF358 | 0.734412 | 3 | 0 | 3 |
| MICA | NADK | 0.73412 | 3 | 0 | 3 |
| MICA | ME3 | 0.731249 | 3 | 0 | 3 |
| MICA | PBXIP1 | 0.728002 | 3 | 0 | 3 |
| MICA | CYP27A1 | 0.725133 | 3 | 0 | 3 |
| MICA | CLIP3 | 0.720821 | 3 | 0 | 3 |
| MICA | CACNA2D1 | 0.715678 | 3 | 0 | 3 |
| MICA | AFF1 | 0.711876 | 3 | 0 | 3 |
| MICA | ZNF583 | 0.710734 | 3 | 0 | 3 |
| MICA | DIXDC1 | 0.705008 | 3 | 0 | 3 |
| MICA | PCDH9 | 0.702983 | 4 | 0 | 4 |
| MICA | EEPD1 | 0.694553 | 3 | 0 | 3 |
| MICA | FIBIN | 0.690912 | 3 | 0 | 3 |
| MICA | FAM160B1 | 0.685083 | 4 | 0 | 4 |
| MICA | NUPR1 | 0.680691 | 3 | 0 | 3 |
| MICA | RARS2 | 0.677008 | 3 | 0 | 3 |
| MICA | ANK2 | 0.673954 | 3 | 0 | 3 |
| MICA | PACSIN2 | 0.67337 | 4 | 0 | 3 |
| MICA | C19orf18 | 0.67207 | 3 | 0 | 3 |
For details and further investigation, click here