| Full name: C-X-C motif chemokine ligand 12 | Alias Symbol: SCYB12|SDF-1a|SDF-1b|PBSF|TLSF-a|TLSF-b|TPAR1 | ||
| Type: protein-coding gene | Cytoband: 10q11.21 | ||
| Entrez ID: 6387 | HGNC ID: HGNC:10672 | Ensembl Gene: ENSG00000107562 | OMIM ID: 600835 |
| Drug and gene relationship at DGIdb | |||
CXCL12 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04062 | Chemokine signaling pathway | |
| hsa04064 | NF-kappa B signaling pathway | |
| hsa04360 | Axon guidance | |
| hsa04670 | Leukocyte transendothelial migration |
Expression of CXCL12:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CXCL12 | 6387 | 209687_at | -3.3038 | 0.0559 | |
| GSE20347 | CXCL12 | 6387 | 209687_at | -1.5404 | 0.0001 | |
| GSE23400 | CXCL12 | 6387 | 203666_at | -0.2037 | 0.0111 | |
| GSE26886 | CXCL12 | 6387 | 203666_at | -0.2865 | 0.2847 | |
| GSE29001 | CXCL12 | 6387 | 209687_at | -1.2680 | 0.0102 | |
| GSE38129 | CXCL12 | 6387 | 209687_at | -2.2860 | 0.0001 | |
| GSE45670 | CXCL12 | 6387 | 209687_at | -5.4010 | 0.0000 | |
| GSE53622 | CXCL12 | 6387 | 102093 | -1.8266 | 0.0000 | |
| GSE53624 | CXCL12 | 6387 | 102093 | -1.5409 | 0.0000 | |
| GSE63941 | CXCL12 | 6387 | 203666_at | -2.5191 | 0.0000 | |
| GSE77861 | CXCL12 | 6387 | 203666_at | -0.0514 | 0.8270 | |
| GSE97050 | CXCL12 | 6387 | A_33_P3712341 | -0.4919 | 0.1421 | |
| SRP007169 | CXCL12 | 6387 | RNAseq | 0.0156 | 0.9818 | |
| SRP008496 | CXCL12 | 6387 | RNAseq | 1.2344 | 0.1065 | |
| SRP064894 | CXCL12 | 6387 | RNAseq | -0.9761 | 0.0293 | |
| SRP133303 | CXCL12 | 6387 | RNAseq | -2.2057 | 0.0000 | |
| SRP159526 | CXCL12 | 6387 | RNAseq | -1.5920 | 0.0006 | |
| SRP193095 | CXCL12 | 6387 | RNAseq | 0.2592 | 0.5358 | |
| SRP219564 | CXCL12 | 6387 | RNAseq | -0.3567 | 0.7334 | |
| TCGA | CXCL12 | 6387 | RNAseq | -0.7685 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 9.
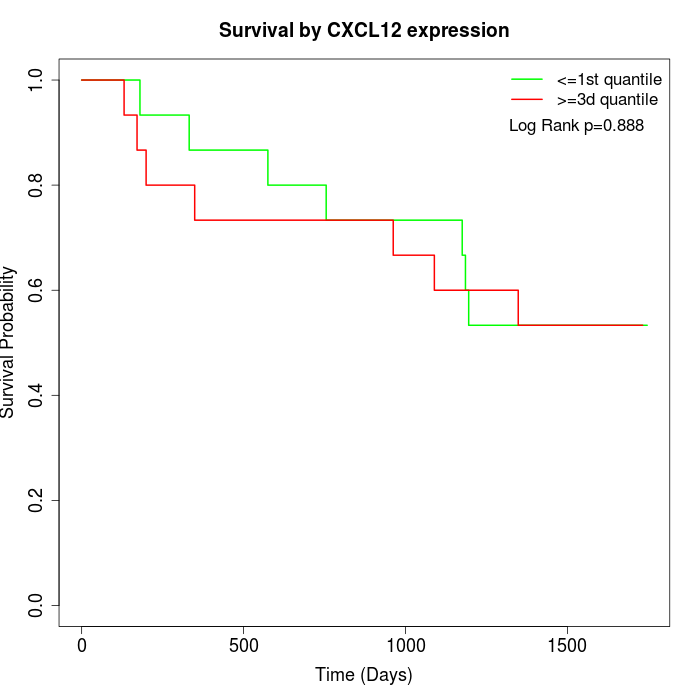
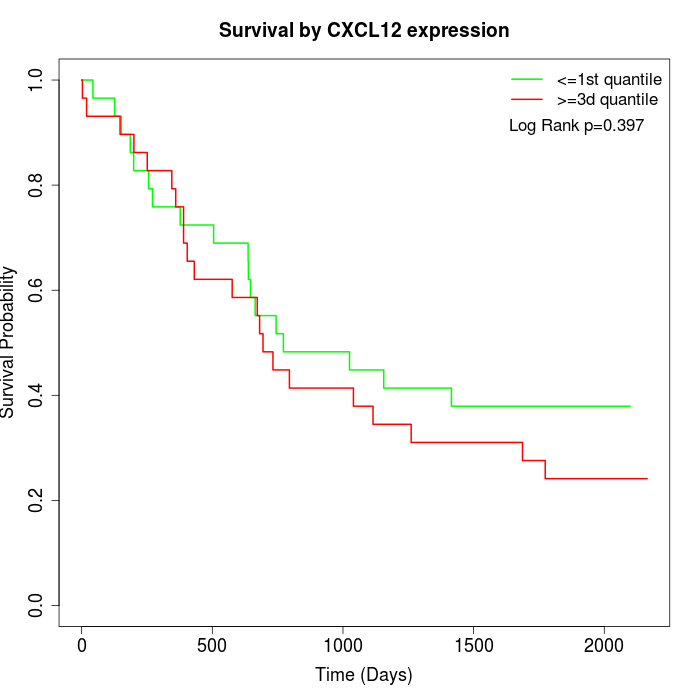
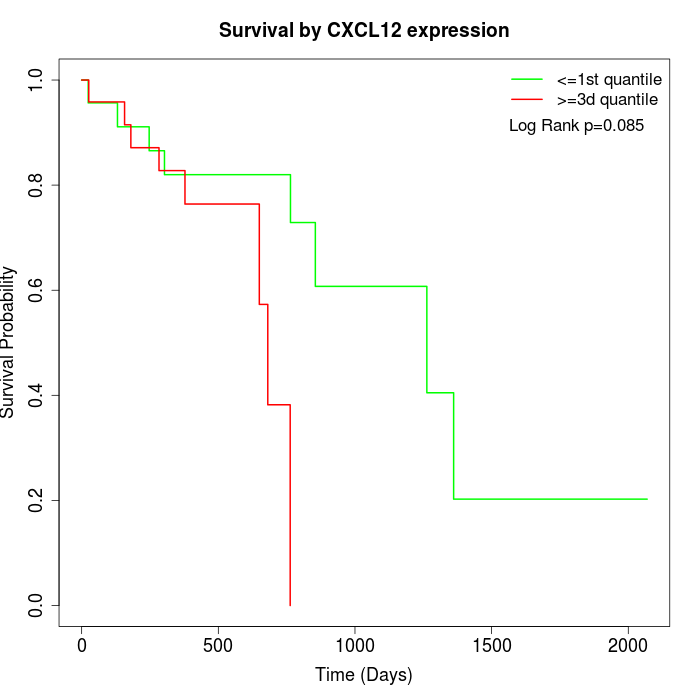
Survival by CXCL12 expression:
Note: Click image to view full size file.
Copy number change of CXCL12:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CXCL12 | 6387 | 3 | 5 | 22 | |
| GSE20123 | CXCL12 | 6387 | 3 | 5 | 22 | |
| GSE43470 | CXCL12 | 6387 | 1 | 6 | 36 | |
| GSE46452 | CXCL12 | 6387 | 0 | 13 | 46 | |
| GSE47630 | CXCL12 | 6387 | 4 | 12 | 24 | |
| GSE54993 | CXCL12 | 6387 | 9 | 0 | 61 | |
| GSE54994 | CXCL12 | 6387 | 2 | 10 | 41 | |
| GSE60625 | CXCL12 | 6387 | 0 | 0 | 11 | |
| GSE74703 | CXCL12 | 6387 | 1 | 3 | 32 | |
| GSE74704 | CXCL12 | 6387 | 2 | 4 | 14 | |
| TCGA | CXCL12 | 6387 | 17 | 19 | 60 |
Total number of gains: 42; Total number of losses: 77; Total Number of normals: 369.
Somatic mutations of CXCL12:
Generating mutation plots.
Highly correlated genes for CXCL12:
Showing top 20/1431 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CXCL12 | CYP21A2 | 0.86194 | 3 | 0 | 3 |
| CXCL12 | FIBIN | 0.853378 | 5 | 0 | 5 |
| CXCL12 | CLEC3B | 0.848157 | 3 | 0 | 3 |
| CXCL12 | LINC01082 | 0.834104 | 5 | 0 | 5 |
| CXCL12 | ZNF132 | 0.832868 | 3 | 0 | 3 |
| CXCL12 | SYNPO2 | 0.82621 | 6 | 0 | 6 |
| CXCL12 | C11orf96 | 0.820754 | 4 | 0 | 4 |
| CXCL12 | ABCA8 | 0.811808 | 11 | 0 | 10 |
| CXCL12 | TMEM100 | 0.797776 | 10 | 0 | 10 |
| CXCL12 | COL14A1 | 0.791121 | 11 | 0 | 10 |
| CXCL12 | ANGPTL1 | 0.789126 | 7 | 0 | 6 |
| CXCL12 | ZBTB47 | 0.788293 | 3 | 0 | 3 |
| CXCL12 | ARHGAP6 | 0.787823 | 11 | 0 | 11 |
| CXCL12 | RCAN2 | 0.785195 | 12 | 0 | 11 |
| CXCL12 | SMOC2 | 0.784708 | 5 | 0 | 5 |
| CXCL12 | COX7A1 | 0.78284 | 10 | 0 | 10 |
| CXCL12 | PGM5-AS1 | 0.781316 | 3 | 0 | 3 |
| CXCL12 | FZD7 | 0.779247 | 3 | 0 | 3 |
| CXCL12 | LRRK2 | 0.778609 | 6 | 0 | 6 |
| CXCL12 | AOC3 | 0.776544 | 11 | 0 | 10 |
For details and further investigation, click here