| Full name: cyclin dependent kinase 15 | Alias Symbol: PFTAIRE2 | ||
| Type: protein-coding gene | Cytoband: 2q33.1 | ||
| Entrez ID: 65061 | HGNC ID: HGNC:14434 | Ensembl Gene: ENSG00000138395 | OMIM ID: 616147 |
| Drug and gene relationship at DGIdb | |||
Expression of CDK15:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CDK15 | 65061 | 239201_at | -0.1820 | 0.3905 | |
| GSE26886 | CDK15 | 65061 | 239201_at | -0.1200 | 0.5261 | |
| GSE45670 | CDK15 | 65061 | 239201_at | -0.4387 | 0.0000 | |
| GSE53622 | CDK15 | 65061 | 35708 | -0.3219 | 0.0001 | |
| GSE53624 | CDK15 | 65061 | 35708 | -0.2154 | 0.0000 | |
| GSE63941 | CDK15 | 65061 | 239201_at | -1.3281 | 0.0051 | |
| GSE77861 | CDK15 | 65061 | 239201_at | -0.1946 | 0.2849 | |
| GSE97050 | CDK15 | 65061 | A_23_P154050 | -0.2018 | 0.3907 | |
| SRP133303 | CDK15 | 65061 | RNAseq | -0.6264 | 0.0109 | |
| TCGA | CDK15 | 65061 | RNAseq | -0.9127 | 0.1273 |
Upregulated datasets: 0; Downregulated datasets: 1.
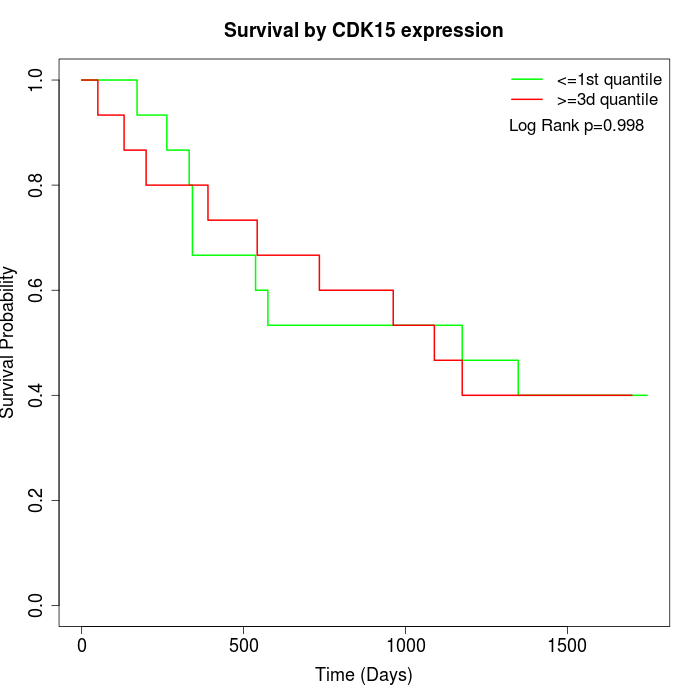
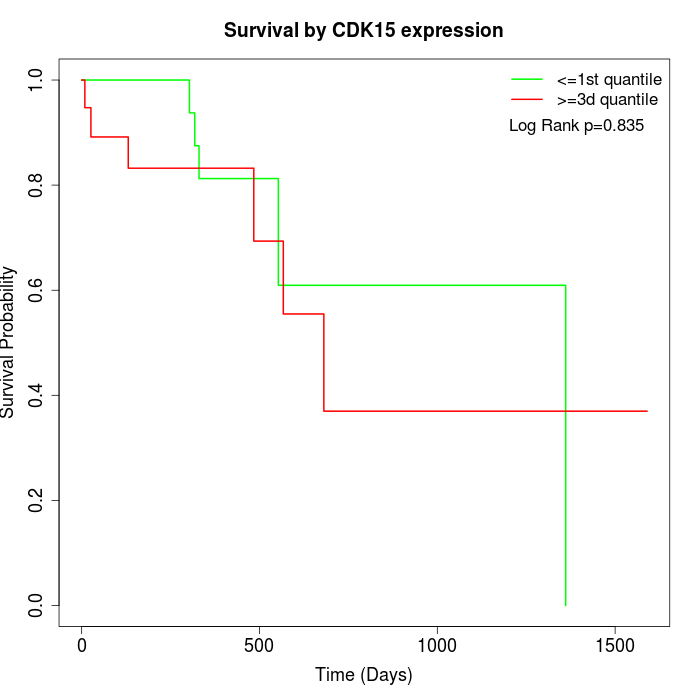
Survival by CDK15 expression:
Note: Click image to view full size file.
Copy number change of CDK15:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CDK15 | 65061 | 6 | 4 | 20 | |
| GSE20123 | CDK15 | 65061 | 6 | 4 | 20 | |
| GSE43470 | CDK15 | 65061 | 1 | 1 | 41 | |
| GSE46452 | CDK15 | 65061 | 1 | 4 | 54 | |
| GSE47630 | CDK15 | 65061 | 4 | 5 | 31 | |
| GSE54993 | CDK15 | 65061 | 0 | 3 | 67 | |
| GSE54994 | CDK15 | 65061 | 10 | 9 | 34 | |
| GSE60625 | CDK15 | 65061 | 0 | 3 | 8 | |
| GSE74703 | CDK15 | 65061 | 1 | 1 | 34 | |
| GSE74704 | CDK15 | 65061 | 3 | 2 | 15 | |
| TCGA | CDK15 | 65061 | 21 | 11 | 64 |
Total number of gains: 53; Total number of losses: 47; Total Number of normals: 388.
Somatic mutations of CDK15:
Generating mutation plots.
Highly correlated genes for CDK15:
Showing top 20/280 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CDK15 | LINC01111 | 0.758213 | 4 | 0 | 3 |
| CDK15 | LINC01279 | 0.748199 | 3 | 0 | 3 |
| CDK15 | TMEM178A | 0.735878 | 3 | 0 | 3 |
| CDK15 | LINC00612 | 0.72401 | 3 | 0 | 3 |
| CDK15 | ZNF781 | 0.718701 | 3 | 0 | 3 |
| CDK15 | GREM2 | 0.716462 | 5 | 0 | 4 |
| CDK15 | LINC01140 | 0.709334 | 3 | 0 | 3 |
| CDK15 | LUZP4 | 0.709233 | 3 | 0 | 3 |
| CDK15 | SLC4A4 | 0.70568 | 4 | 0 | 4 |
| CDK15 | LINC01082 | 0.703227 | 3 | 0 | 3 |
| CDK15 | ZNF471 | 0.701396 | 3 | 0 | 3 |
| CDK15 | CCDC96 | 0.701253 | 3 | 0 | 3 |
| CDK15 | LMO3 | 0.698398 | 3 | 0 | 3 |
| CDK15 | WNT6 | 0.697139 | 4 | 0 | 4 |
| CDK15 | MTFR1L | 0.689802 | 3 | 0 | 3 |
| CDK15 | OR52A1 | 0.686668 | 3 | 0 | 3 |
| CDK15 | ASPA | 0.682997 | 5 | 0 | 4 |
| CDK15 | MAGI2-AS3 | 0.682677 | 5 | 0 | 4 |
| CDK15 | PDE5A | 0.682242 | 6 | 0 | 5 |
| CDK15 | HSPB2 | 0.681665 | 4 | 0 | 3 |
For details and further investigation, click here