| Full name: NOBOX oogenesis homeobox | Alias Symbol: OG2|Og2x | ||
| Type: protein-coding gene | Cytoband: 7q35 | ||
| Entrez ID: 135935 | HGNC ID: HGNC:22448 | Ensembl Gene: ENSG00000106410 | OMIM ID: 610934 |
| Drug and gene relationship at DGIdb | |||
Expression of NOBOX:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NOBOX | 135935 | 234519_at | 0.0264 | 0.9284 | |
| GSE26886 | NOBOX | 135935 | 234519_at | -0.1009 | 0.4822 | |
| GSE45670 | NOBOX | 135935 | 234519_at | 0.0923 | 0.3254 | |
| GSE53622 | NOBOX | 135935 | 60205 | -0.2221 | 0.0134 | |
| GSE53624 | NOBOX | 135935 | 60205 | -0.5183 | 0.0000 | |
| GSE63941 | NOBOX | 135935 | 234519_at | 0.1941 | 0.2660 | |
| GSE77861 | NOBOX | 135935 | 234519_at | -0.1902 | 0.2130 | |
| GSE97050 | NOBOX | 135935 | A_33_P3236310 | 0.0621 | 0.8020 | |
| TCGA | NOBOX | 135935 | RNAseq | 2.5199 | 0.4286 |
Upregulated datasets: 0; Downregulated datasets: 0.
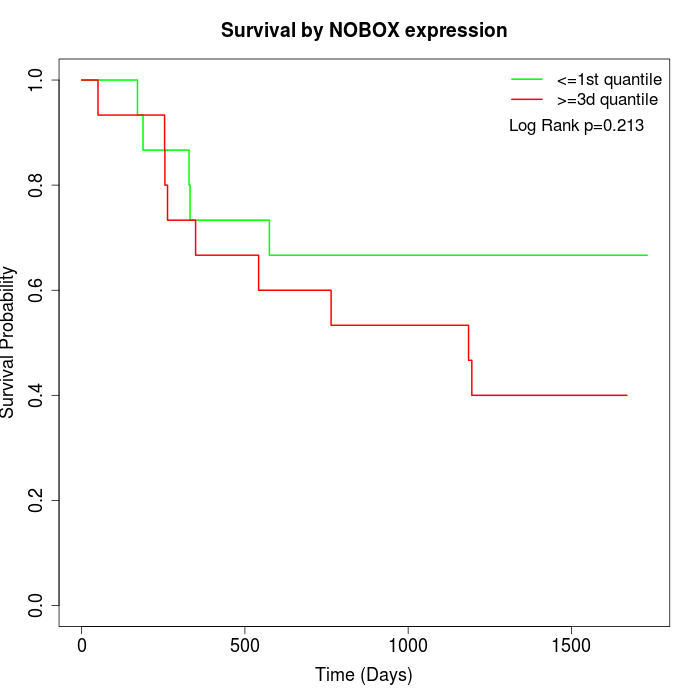
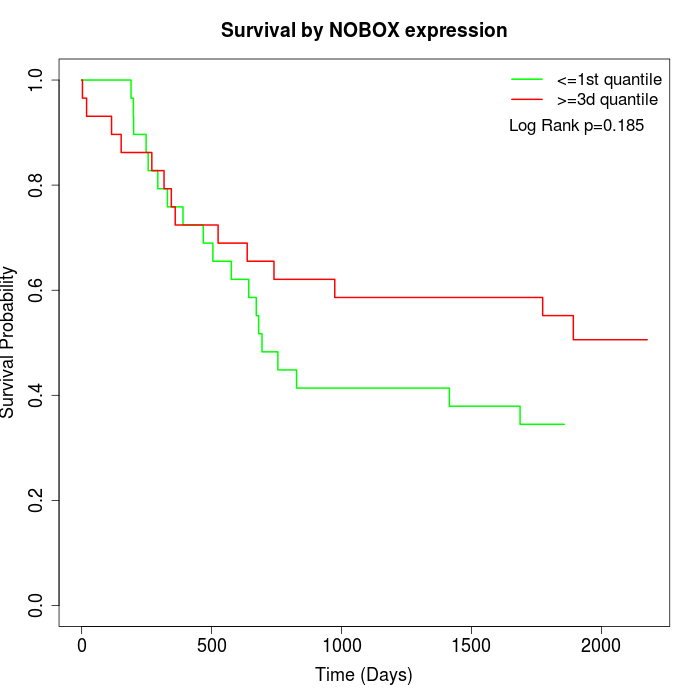
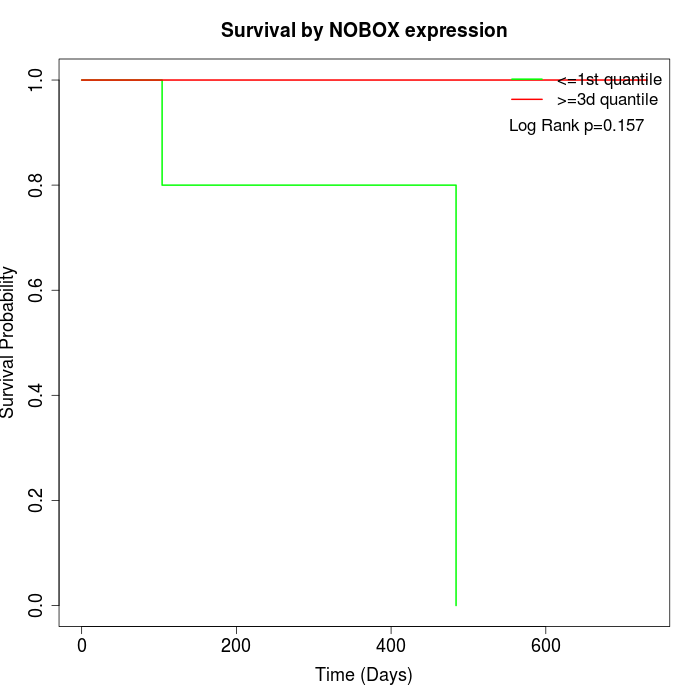
Survival by NOBOX expression:
Note: Click image to view full size file.
Copy number change of NOBOX:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NOBOX | 135935 | 3 | 2 | 25 | |
| GSE20123 | NOBOX | 135935 | 3 | 2 | 25 | |
| GSE43470 | NOBOX | 135935 | 3 | 5 | 35 | |
| GSE46452 | NOBOX | 135935 | 7 | 2 | 50 | |
| GSE47630 | NOBOX | 135935 | 6 | 8 | 26 | |
| GSE54993 | NOBOX | 135935 | 3 | 4 | 63 | |
| GSE54994 | NOBOX | 135935 | 5 | 8 | 40 | |
| GSE60625 | NOBOX | 135935 | 0 | 0 | 11 | |
| GSE74703 | NOBOX | 135935 | 2 | 5 | 29 | |
| GSE74704 | NOBOX | 135935 | 1 | 2 | 17 | |
| TCGA | NOBOX | 135935 | 27 | 27 | 42 |
Total number of gains: 60; Total number of losses: 65; Total Number of normals: 363.
Somatic mutations of NOBOX:
Generating mutation plots.
Highly correlated genes for NOBOX:
Showing top 20/522 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NOBOX | KRTAP6-1 | 0.839216 | 3 | 0 | 3 |
| NOBOX | YY2 | 0.830028 | 3 | 0 | 3 |
| NOBOX | CELF5 | 0.817283 | 3 | 0 | 3 |
| NOBOX | ANKRD33B | 0.815658 | 3 | 0 | 3 |
| NOBOX | RNF180 | 0.809519 | 3 | 0 | 3 |
| NOBOX | OR13G1 | 0.80692 | 3 | 0 | 3 |
| NOBOX | ACSM5 | 0.802858 | 3 | 0 | 3 |
| NOBOX | GDF7 | 0.802415 | 3 | 0 | 3 |
| NOBOX | OR10AD1 | 0.78832 | 3 | 0 | 3 |
| NOBOX | PRSS38 | 0.787328 | 3 | 0 | 3 |
| NOBOX | CLRN2 | 0.78046 | 3 | 0 | 3 |
| NOBOX | CD164L2 | 0.779536 | 3 | 0 | 3 |
| NOBOX | MRGPRE | 0.777098 | 3 | 0 | 3 |
| NOBOX | KRT77 | 0.769279 | 3 | 0 | 3 |
| NOBOX | MFSD2B | 0.764312 | 3 | 0 | 3 |
| NOBOX | TTC21A | 0.760529 | 3 | 0 | 3 |
| NOBOX | MEX3D | 0.759466 | 3 | 0 | 3 |
| NOBOX | PLA2G2C | 0.758457 | 3 | 0 | 3 |
| NOBOX | OTOG | 0.755951 | 3 | 0 | 3 |
| NOBOX | CNGB1 | 0.754947 | 3 | 0 | 3 |
For details and further investigation, click here