| Full name: t-complex-associated-testis-expressed 1 | Alias Symbol: D6S46|MGC33600|FAP155|DRC5 | ||
| Type: protein-coding gene | Cytoband: 6p21.1 | ||
| Entrez ID: 202500 | HGNC ID: HGNC:11693 | Ensembl Gene: ENSG00000146221 | OMIM ID: 186975 |
| Drug and gene relationship at DGIdb | |||
Expression of TCTE1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TCTE1 | 202500 | 237280_at | 0.0338 | 0.9030 | |
| GSE26886 | TCTE1 | 202500 | 237280_at | 0.2961 | 0.0163 | |
| GSE45670 | TCTE1 | 202500 | 237280_at | 0.0106 | 0.9137 | |
| GSE53622 | TCTE1 | 202500 | 98691 | 0.0880 | 0.1373 | |
| GSE53624 | TCTE1 | 202500 | 98691 | 0.2106 | 0.0215 | |
| GSE63941 | TCTE1 | 202500 | 237280_at | 0.1450 | 0.3406 | |
| GSE77861 | TCTE1 | 202500 | 237280_at | -0.0378 | 0.7932 | |
| GSE97050 | TCTE1 | 202500 | A_32_P203099 | 0.0324 | 0.9179 | |
| SRP133303 | TCTE1 | 202500 | RNAseq | 0.0165 | 0.9433 | |
| TCGA | TCTE1 | 202500 | RNAseq | 0.6524 | 0.3436 |
Upregulated datasets: 0; Downregulated datasets: 0.
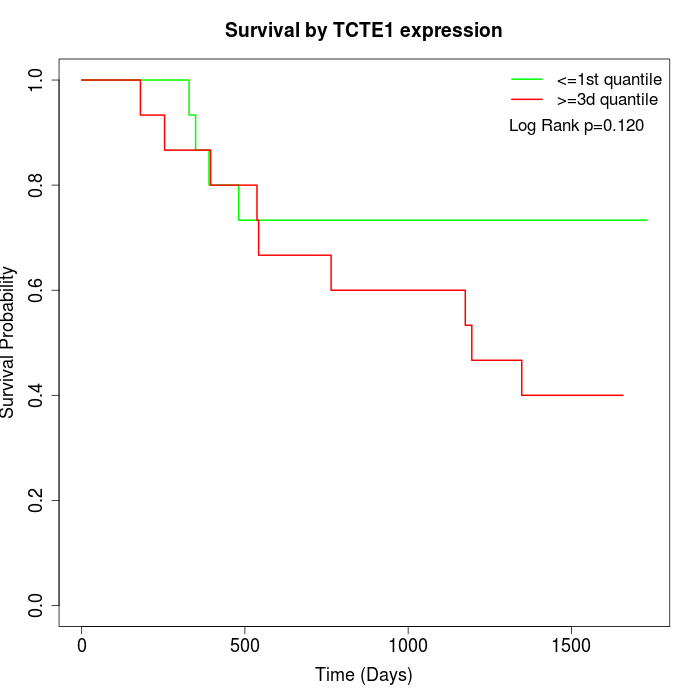
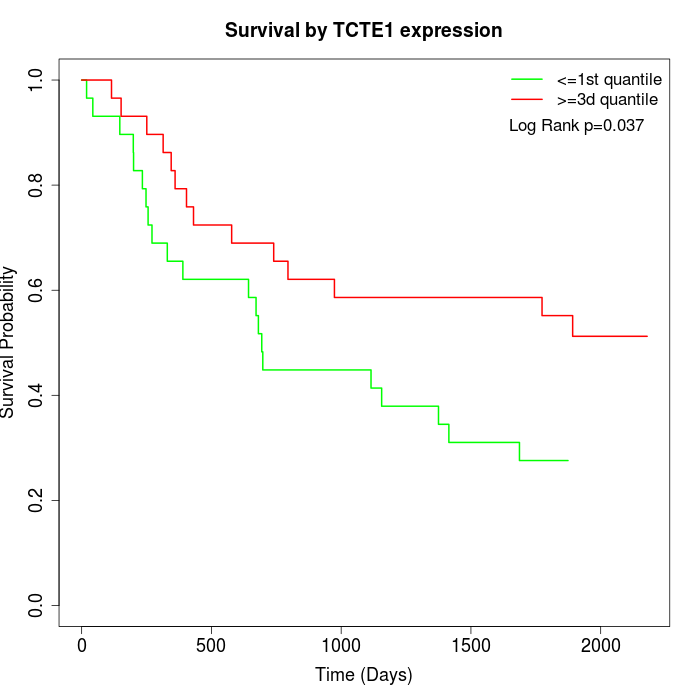
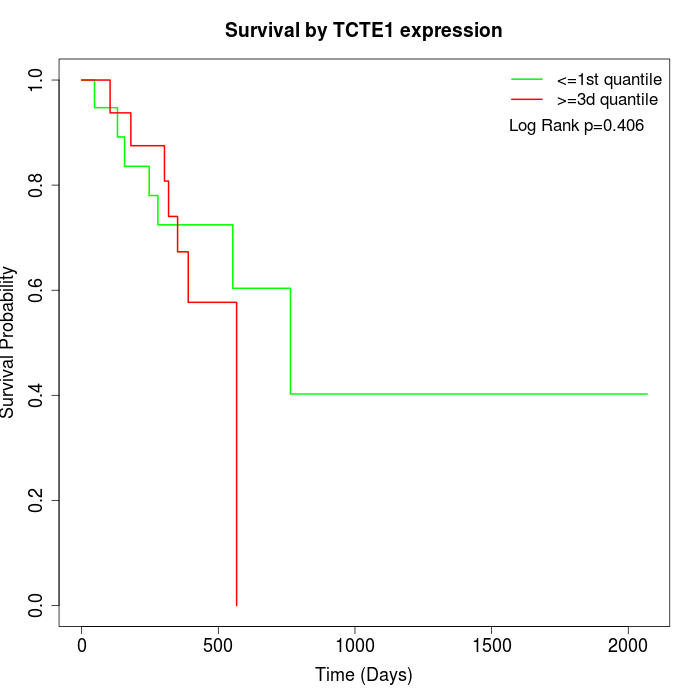
Survival by TCTE1 expression:
Note: Click image to view full size file.
Copy number change of TCTE1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TCTE1 | 202500 | 7 | 1 | 22 | |
| GSE20123 | TCTE1 | 202500 | 7 | 1 | 22 | |
| GSE43470 | TCTE1 | 202500 | 6 | 0 | 37 | |
| GSE46452 | TCTE1 | 202500 | 3 | 9 | 47 | |
| GSE47630 | TCTE1 | 202500 | 8 | 4 | 28 | |
| GSE54993 | TCTE1 | 202500 | 3 | 2 | 65 | |
| GSE54994 | TCTE1 | 202500 | 8 | 3 | 42 | |
| GSE60625 | TCTE1 | 202500 | 0 | 3 | 8 | |
| GSE74703 | TCTE1 | 202500 | 6 | 0 | 30 | |
| GSE74704 | TCTE1 | 202500 | 3 | 1 | 16 | |
| TCGA | TCTE1 | 202500 | 21 | 14 | 61 |
Total number of gains: 72; Total number of losses: 38; Total Number of normals: 378.
Somatic mutations of TCTE1:
Generating mutation plots.
Highly correlated genes for TCTE1:
Showing top 20/431 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TCTE1 | STRC | 0.858315 | 3 | 0 | 3 |
| TCTE1 | GRIK4 | 0.841339 | 3 | 0 | 3 |
| TCTE1 | NKX6-3 | 0.817243 | 3 | 0 | 3 |
| TCTE1 | DCDC1 | 0.813516 | 3 | 0 | 3 |
| TCTE1 | CELF5 | 0.811075 | 3 | 0 | 3 |
| TCTE1 | C2CD4D | 0.806889 | 3 | 0 | 3 |
| TCTE1 | PLA2G2C | 0.80618 | 3 | 0 | 3 |
| TCTE1 | C9orf153 | 0.798669 | 3 | 0 | 3 |
| TCTE1 | C19orf71 | 0.793525 | 3 | 0 | 3 |
| TCTE1 | GHSR | 0.785 | 3 | 0 | 3 |
| TCTE1 | PRDM13 | 0.783437 | 3 | 0 | 3 |
| TCTE1 | KRTAP10-12 | 0.782732 | 3 | 0 | 3 |
| TCTE1 | FBXO17 | 0.774984 | 3 | 0 | 3 |
| TCTE1 | YY2 | 0.773801 | 3 | 0 | 3 |
| TCTE1 | SNED1 | 0.770876 | 3 | 0 | 3 |
| TCTE1 | MRGPRG | 0.767555 | 3 | 0 | 3 |
| TCTE1 | SERPINA10 | 0.763009 | 4 | 0 | 4 |
| TCTE1 | LRTM1 | 0.76148 | 3 | 0 | 3 |
| TCTE1 | TMEM105 | 0.760097 | 4 | 0 | 4 |
| TCTE1 | OR2T27 | 0.759838 | 3 | 0 | 3 |
For details and further investigation, click here