| Full name: colorectal cancer associated 2 | Alias Symbol: CASC13 | ||
| Type: protein-coding gene | Cytoband: 11q23.1 | ||
| Entrez ID: 120376 | HGNC ID: HGNC:26978 | Ensembl Gene: ENSG00000214290 | OMIM ID: 615694 |
| Drug and gene relationship at DGIdb | |||
Expression of COLCA2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | COLCA2 | 120376 | 228338_at | -0.4007 | 0.6951 | |
| GSE26886 | COLCA2 | 120376 | 228338_at | -0.2234 | 0.7661 | |
| GSE45670 | COLCA2 | 120376 | 228338_at | 0.0167 | 0.9784 | |
| GSE53622 | COLCA2 | 120376 | 28092 | -0.4626 | 0.1047 | |
| GSE53624 | COLCA2 | 120376 | 28092 | -0.4261 | 0.0406 | |
| GSE63941 | COLCA2 | 120376 | 228338_at | 2.1876 | 0.2630 | |
| GSE77861 | COLCA2 | 120376 | 228338_at | 0.1538 | 0.8019 | |
| SRP064894 | COLCA2 | 120376 | RNAseq | 0.5849 | 0.2115 | |
| SRP133303 | COLCA2 | 120376 | RNAseq | -0.3079 | 0.4507 | |
| SRP193095 | COLCA2 | 120376 | RNAseq | -0.1685 | 0.6338 | |
| SRP219564 | COLCA2 | 120376 | RNAseq | 1.0899 | 0.1639 |
Upregulated datasets: 0; Downregulated datasets: 0.
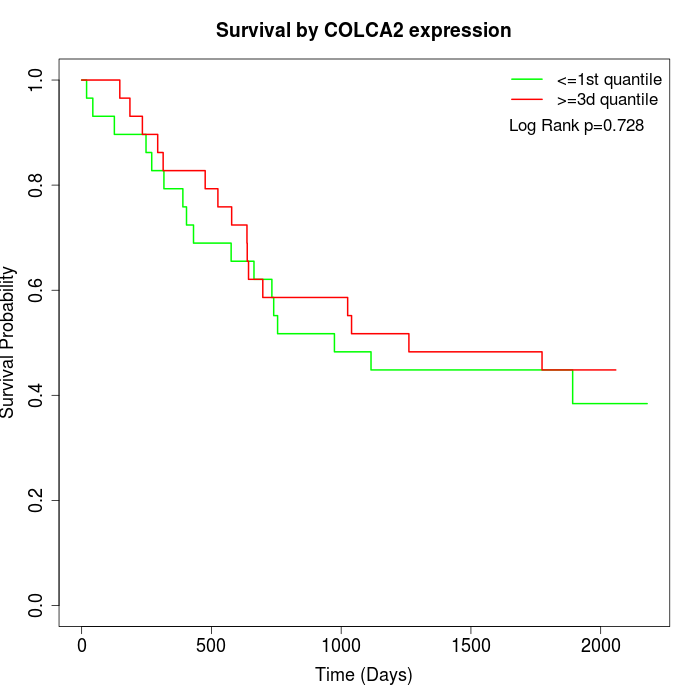
Survival by COLCA2 expression:
Note: Click image to view full size file.
Copy number change of COLCA2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | COLCA2 | 120376 | 0 | 12 | 18 | |
| GSE20123 | COLCA2 | 120376 | 0 | 12 | 18 | |
| GSE43470 | COLCA2 | 120376 | 2 | 6 | 35 | |
| GSE46452 | COLCA2 | 120376 | 3 | 26 | 30 | |
| GSE47630 | COLCA2 | 120376 | 2 | 19 | 19 | |
| GSE54993 | COLCA2 | 120376 | 10 | 0 | 60 | |
| GSE54994 | COLCA2 | 120376 | 5 | 19 | 29 | |
| GSE60625 | COLCA2 | 120376 | 0 | 3 | 8 | |
| GSE74703 | COLCA2 | 120376 | 1 | 4 | 31 | |
| GSE74704 | COLCA2 | 120376 | 0 | 8 | 12 |
Total number of gains: 23; Total number of losses: 109; Total Number of normals: 260.
Somatic mutations of COLCA2:
Generating mutation plots.
Highly correlated genes for COLCA2:
Showing top 20/37 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| COLCA2 | ZBED5 | 0.695294 | 3 | 0 | 3 |
| COLCA2 | RASSF6 | 0.683599 | 3 | 0 | 3 |
| COLCA2 | BET1L | 0.600268 | 3 | 0 | 3 |
| COLCA2 | TSPAN3 | 0.588618 | 4 | 0 | 3 |
| COLCA2 | TMEM62 | 0.583925 | 4 | 0 | 3 |
| COLCA2 | SCNN1A | 0.583444 | 3 | 0 | 3 |
| COLCA2 | MECOM | 0.580758 | 6 | 0 | 4 |
| COLCA2 | ANXA11 | 0.580265 | 3 | 0 | 3 |
| COLCA2 | TRIM68 | 0.580122 | 5 | 0 | 3 |
| COLCA2 | ALDH1A1 | 0.578083 | 4 | 0 | 3 |
| COLCA2 | ILDR1 | 0.574022 | 4 | 0 | 3 |
| COLCA2 | LDOC1 | 0.573701 | 4 | 0 | 3 |
| COLCA2 | GCNT2 | 0.56583 | 6 | 0 | 4 |
| COLCA2 | KIAA1671 | 0.563202 | 4 | 0 | 3 |
| COLCA2 | LINC01003 | 0.561577 | 4 | 0 | 3 |
| COLCA2 | MPPED2 | 0.561409 | 3 | 0 | 3 |
| COLCA2 | PLXNA2 | 0.560948 | 3 | 0 | 3 |
| COLCA2 | C11orf1 | 0.560587 | 3 | 0 | 3 |
| COLCA2 | CLDN8 | 0.558378 | 7 | 0 | 4 |
| COLCA2 | EPHX1 | 0.554591 | 6 | 0 | 4 |
For details and further investigation, click here