| Full name: tripartite motif containing 68 | Alias Symbol: SS-56|FLJ10369 | ||
| Type: protein-coding gene | Cytoband: 11p15.4 | ||
| Entrez ID: 55128 | HGNC ID: HGNC:21161 | Ensembl Gene: ENSG00000167333 | OMIM ID: 613184 |
| Drug and gene relationship at DGIdb | |||
Expression of TRIM68:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TRIM68 | 55128 | 219405_at | -0.2576 | 0.6029 | |
| GSE20347 | TRIM68 | 55128 | 219405_at | 0.1930 | 0.2279 | |
| GSE23400 | TRIM68 | 55128 | 219405_at | 0.1947 | 0.0019 | |
| GSE26886 | TRIM68 | 55128 | 219405_at | 0.3048 | 0.4099 | |
| GSE29001 | TRIM68 | 55128 | 219405_at | 0.3012 | 0.2144 | |
| GSE38129 | TRIM68 | 55128 | 219405_at | -0.0400 | 0.8458 | |
| GSE45670 | TRIM68 | 55128 | 219405_at | 0.1343 | 0.4007 | |
| GSE53622 | TRIM68 | 55128 | 59042 | -0.2393 | 0.0511 | |
| GSE53624 | TRIM68 | 55128 | 59042 | 0.2006 | 0.0476 | |
| GSE63941 | TRIM68 | 55128 | 219405_at | -0.6946 | 0.2772 | |
| GSE77861 | TRIM68 | 55128 | 219405_at | 0.0214 | 0.9413 | |
| GSE97050 | TRIM68 | 55128 | A_23_P2097 | 0.0705 | 0.8017 | |
| SRP007169 | TRIM68 | 55128 | RNAseq | 0.1251 | 0.7835 | |
| SRP008496 | TRIM68 | 55128 | RNAseq | 0.6796 | 0.0402 | |
| SRP064894 | TRIM68 | 55128 | RNAseq | 0.1550 | 0.3065 | |
| SRP133303 | TRIM68 | 55128 | RNAseq | 0.0768 | 0.5950 | |
| SRP159526 | TRIM68 | 55128 | RNAseq | -0.1482 | 0.6924 | |
| SRP193095 | TRIM68 | 55128 | RNAseq | -0.1440 | 0.2796 | |
| SRP219564 | TRIM68 | 55128 | RNAseq | -0.1108 | 0.7686 | |
| TCGA | TRIM68 | 55128 | RNAseq | -0.3196 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 0.
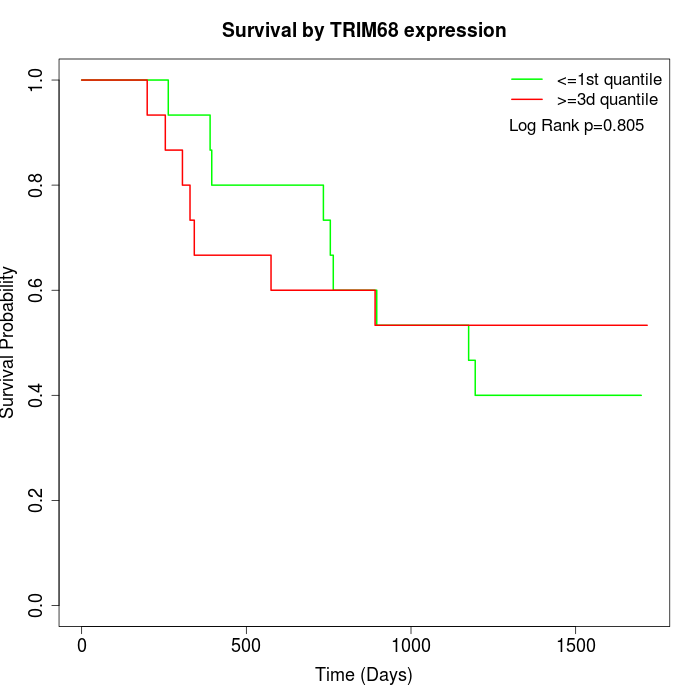
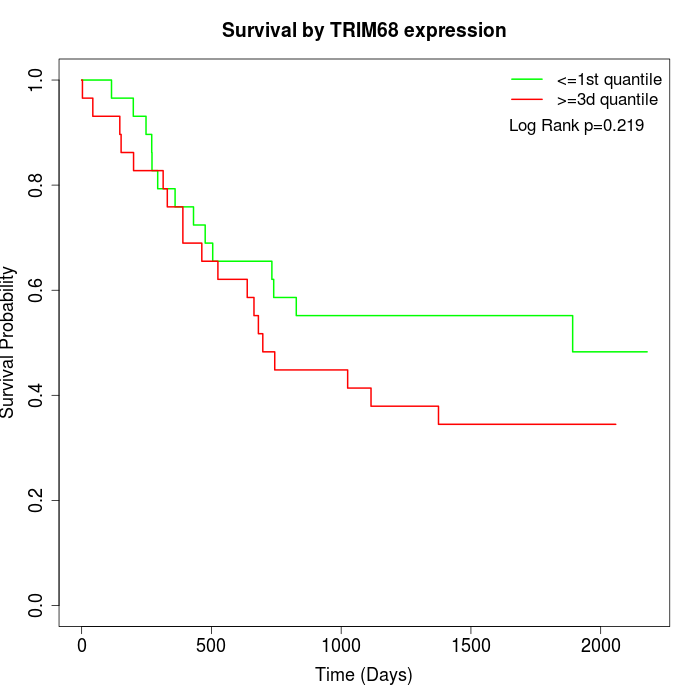
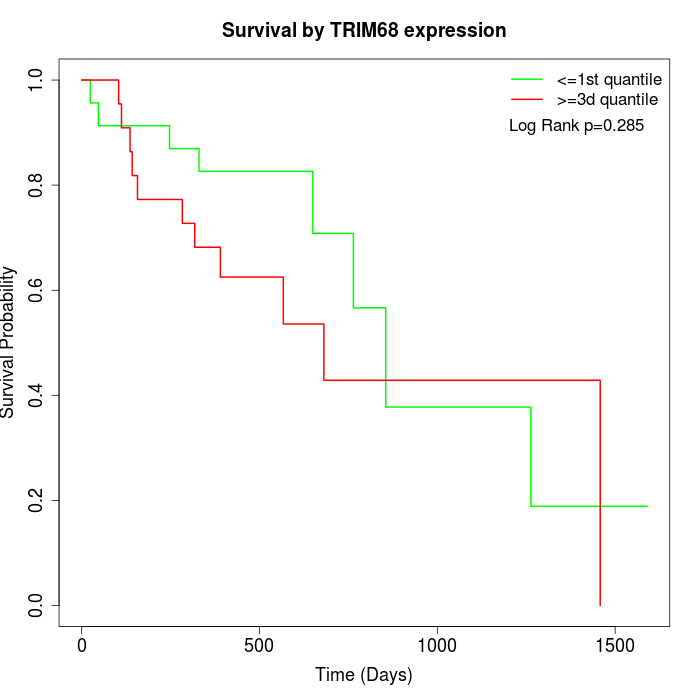
Survival by TRIM68 expression:
Note: Click image to view full size file.
Copy number change of TRIM68:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TRIM68 | 55128 | 1 | 12 | 17 | |
| GSE20123 | TRIM68 | 55128 | 1 | 12 | 17 | |
| GSE43470 | TRIM68 | 55128 | 1 | 8 | 34 | |
| GSE46452 | TRIM68 | 55128 | 7 | 8 | 44 | |
| GSE47630 | TRIM68 | 55128 | 3 | 13 | 24 | |
| GSE54993 | TRIM68 | 55128 | 3 | 1 | 66 | |
| GSE54994 | TRIM68 | 55128 | 1 | 13 | 39 | |
| GSE60625 | TRIM68 | 55128 | 0 | 0 | 11 | |
| GSE74703 | TRIM68 | 55128 | 1 | 6 | 29 | |
| GSE74704 | TRIM68 | 55128 | 0 | 8 | 12 | |
| TCGA | TRIM68 | 55128 | 8 | 39 | 49 |
Total number of gains: 26; Total number of losses: 120; Total Number of normals: 342.
Somatic mutations of TRIM68:
Generating mutation plots.
Highly correlated genes for TRIM68:
Showing top 20/341 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TRIM68 | KAT5 | 0.762741 | 3 | 0 | 3 |
| TRIM68 | ASB8 | 0.747989 | 3 | 0 | 3 |
| TRIM68 | CCDC97 | 0.709732 | 3 | 0 | 3 |
| TRIM68 | GLUD1 | 0.699628 | 3 | 0 | 3 |
| TRIM68 | PUS7L | 0.698028 | 3 | 0 | 3 |
| TRIM68 | SRSF8 | 0.695491 | 3 | 0 | 3 |
| TRIM68 | HIP1 | 0.682815 | 3 | 0 | 3 |
| TRIM68 | ZBED5 | 0.681245 | 7 | 0 | 5 |
| TRIM68 | NCOR1 | 0.675768 | 3 | 0 | 3 |
| TRIM68 | PNN | 0.675071 | 3 | 0 | 3 |
| TRIM68 | SYT17 | 0.671494 | 3 | 0 | 3 |
| TRIM68 | ZNF740 | 0.671491 | 3 | 0 | 3 |
| TRIM68 | NHS | 0.670747 | 3 | 0 | 3 |
| TRIM68 | LRRK1 | 0.670704 | 3 | 0 | 3 |
| TRIM68 | OSCP1 | 0.670548 | 4 | 0 | 3 |
| TRIM68 | ZNF627 | 0.669493 | 3 | 0 | 3 |
| TRIM68 | KLHL15 | 0.667662 | 3 | 0 | 3 |
| TRIM68 | DNALI1 | 0.662999 | 3 | 0 | 3 |
| TRIM68 | ZNF343 | 0.662544 | 3 | 0 | 3 |
| TRIM68 | FAM102B | 0.662514 | 3 | 0 | 3 |
For details and further investigation, click here