| Full name: epoxide hydrolase 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 1q42.12 | ||
| Entrez ID: 2052 | HGNC ID: HGNC:3401 | Ensembl Gene: ENSG00000143819 | OMIM ID: 132810 |
| Drug and gene relationship at DGIdb | |||
Expression of EPHX1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | EPHX1 | 2052 | 202017_at | -0.2230 | 0.7208 | |
| GSE20347 | EPHX1 | 2052 | 202017_at | -0.4963 | 0.0881 | |
| GSE23400 | EPHX1 | 2052 | 202017_at | -0.2332 | 0.1651 | |
| GSE26886 | EPHX1 | 2052 | 202017_at | -0.2270 | 0.5249 | |
| GSE29001 | EPHX1 | 2052 | 202017_at | -0.4456 | 0.0584 | |
| GSE38129 | EPHX1 | 2052 | 202017_at | -0.3098 | 0.2689 | |
| GSE45670 | EPHX1 | 2052 | 202017_at | -0.3979 | 0.1819 | |
| GSE53622 | EPHX1 | 2052 | 21665 | -0.9231 | 0.0000 | |
| GSE53624 | EPHX1 | 2052 | 21665 | -0.7482 | 0.0000 | |
| GSE63941 | EPHX1 | 2052 | 202017_at | 2.6730 | 0.0384 | |
| GSE77861 | EPHX1 | 2052 | 202017_at | -0.1923 | 0.6478 | |
| GSE97050 | EPHX1 | 2052 | A_23_P34537 | -0.3757 | 0.4850 | |
| SRP007169 | EPHX1 | 2052 | RNAseq | -0.9469 | 0.0363 | |
| SRP008496 | EPHX1 | 2052 | RNAseq | -0.8083 | 0.0528 | |
| SRP064894 | EPHX1 | 2052 | RNAseq | -0.6740 | 0.0154 | |
| SRP133303 | EPHX1 | 2052 | RNAseq | -1.1210 | 0.0000 | |
| SRP159526 | EPHX1 | 2052 | RNAseq | 0.4173 | 0.5037 | |
| SRP193095 | EPHX1 | 2052 | RNAseq | -0.1990 | 0.4507 | |
| SRP219564 | EPHX1 | 2052 | RNAseq | -0.1900 | 0.6936 | |
| TCGA | EPHX1 | 2052 | RNAseq | 0.0470 | 0.6398 |
Upregulated datasets: 1; Downregulated datasets: 1.
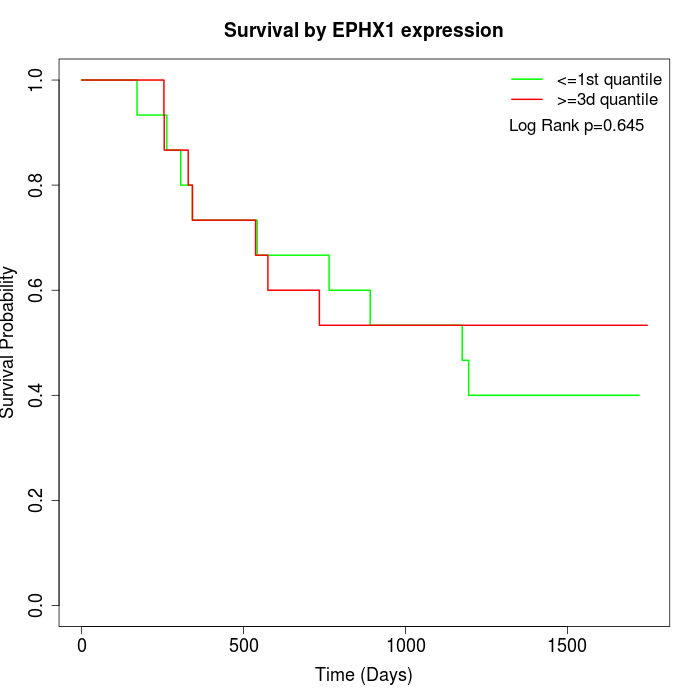
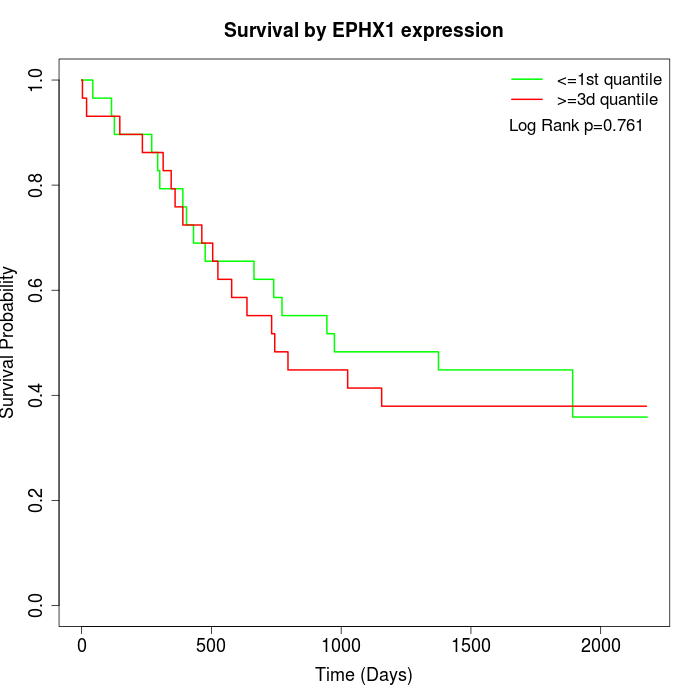
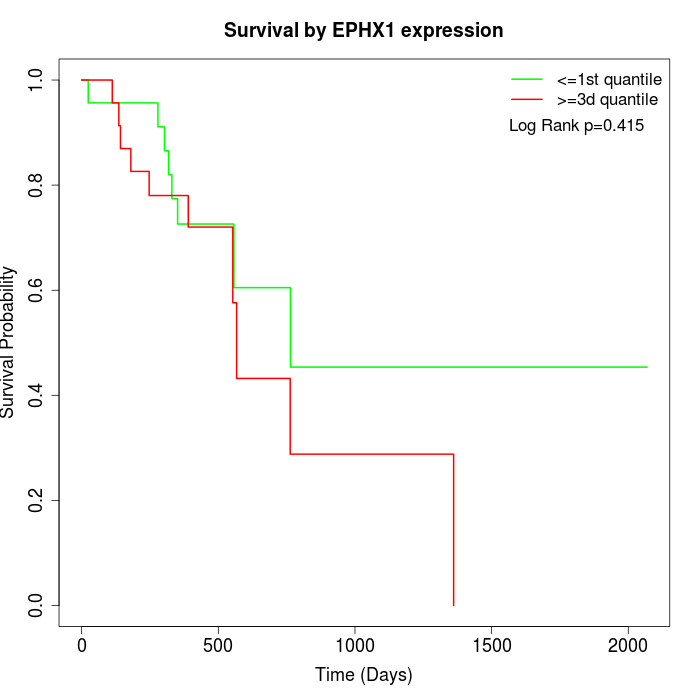
Survival by EPHX1 expression:
Note: Click image to view full size file.
Copy number change of EPHX1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | EPHX1 | 2052 | 11 | 0 | 19 | |
| GSE20123 | EPHX1 | 2052 | 11 | 0 | 19 | |
| GSE43470 | EPHX1 | 2052 | 7 | 1 | 35 | |
| GSE46452 | EPHX1 | 2052 | 3 | 1 | 55 | |
| GSE47630 | EPHX1 | 2052 | 15 | 0 | 25 | |
| GSE54993 | EPHX1 | 2052 | 0 | 6 | 64 | |
| GSE54994 | EPHX1 | 2052 | 16 | 0 | 37 | |
| GSE60625 | EPHX1 | 2052 | 0 | 0 | 11 | |
| GSE74703 | EPHX1 | 2052 | 7 | 1 | 28 | |
| GSE74704 | EPHX1 | 2052 | 5 | 0 | 15 | |
| TCGA | EPHX1 | 2052 | 46 | 3 | 47 |
Total number of gains: 121; Total number of losses: 12; Total Number of normals: 355.
Somatic mutations of EPHX1:
Generating mutation plots.
Highly correlated genes for EPHX1:
Showing top 20/350 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| EPHX1 | MPV17L | 0.733864 | 3 | 0 | 3 |
| EPHX1 | ZNF213 | 0.711554 | 3 | 0 | 3 |
| EPHX1 | RBL2 | 0.705908 | 3 | 0 | 3 |
| EPHX1 | RAB35 | 0.699497 | 3 | 0 | 3 |
| EPHX1 | KLHDC3 | 0.69302 | 3 | 0 | 3 |
| EPHX1 | CCNDBP1 | 0.687666 | 3 | 0 | 3 |
| EPHX1 | TMCO6 | 0.677923 | 3 | 0 | 3 |
| EPHX1 | KAT5 | 0.662847 | 5 | 0 | 4 |
| EPHX1 | CYP4F3 | 0.6613 | 11 | 0 | 10 |
| EPHX1 | DICER1 | 0.656504 | 3 | 0 | 3 |
| EPHX1 | AK3 | 0.654776 | 3 | 0 | 3 |
| EPHX1 | ABCC1 | 0.652138 | 4 | 0 | 3 |
| EPHX1 | PMF1 | 0.648954 | 5 | 0 | 4 |
| EPHX1 | AAAS | 0.648704 | 4 | 0 | 4 |
| EPHX1 | ZNF414 | 0.645766 | 4 | 0 | 3 |
| EPHX1 | LOXL4 | 0.645379 | 3 | 0 | 3 |
| EPHX1 | MAP2K5 | 0.644255 | 3 | 0 | 3 |
| EPHX1 | RGS7BP | 0.639635 | 3 | 0 | 3 |
| EPHX1 | TAF10 | 0.63955 | 5 | 0 | 3 |
| EPHX1 | PTGR1 | 0.636632 | 7 | 0 | 6 |
For details and further investigation, click here