| Full name: plexin A2 | Alias Symbol: OCT|FLJ11751|FLJ30634|KIAA0463 | ||
| Type: protein-coding gene | Cytoband: 1q32.2 | ||
| Entrez ID: 5362 | HGNC ID: HGNC:9100 | Ensembl Gene: ENSG00000076356 | OMIM ID: 601054 |
| Drug and gene relationship at DGIdb | |||
PLXNA2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04360 | Axon guidance |
Expression of PLXNA2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PLXNA2 | 5362 | 213030_s_at | 0.1617 | 0.8838 | |
| GSE20347 | PLXNA2 | 5362 | 213030_s_at | -1.0816 | 0.0001 | |
| GSE23400 | PLXNA2 | 5362 | 213030_s_at | -0.5009 | 0.0000 | |
| GSE26886 | PLXNA2 | 5362 | 227032_at | -1.3662 | 0.0000 | |
| GSE29001 | PLXNA2 | 5362 | 213030_s_at | -0.6650 | 0.0367 | |
| GSE38129 | PLXNA2 | 5362 | 213030_s_at | -0.6499 | 0.0078 | |
| GSE45670 | PLXNA2 | 5362 | 213030_s_at | -0.2837 | 0.1648 | |
| GSE53622 | PLXNA2 | 5362 | 34726 | -1.5492 | 0.0000 | |
| GSE53624 | PLXNA2 | 5362 | 34726 | -1.7084 | 0.0000 | |
| GSE63941 | PLXNA2 | 5362 | 213030_s_at | 1.1694 | 0.1821 | |
| GSE77861 | PLXNA2 | 5362 | 213030_s_at | -0.0954 | 0.7668 | |
| GSE97050 | PLXNA2 | 5362 | A_23_P307544 | -0.3424 | 0.1870 | |
| SRP007169 | PLXNA2 | 5362 | RNAseq | -0.8066 | 0.2419 | |
| SRP008496 | PLXNA2 | 5362 | RNAseq | -0.9365 | 0.0198 | |
| SRP064894 | PLXNA2 | 5362 | RNAseq | -0.8751 | 0.0007 | |
| SRP133303 | PLXNA2 | 5362 | RNAseq | -0.8812 | 0.0000 | |
| SRP159526 | PLXNA2 | 5362 | RNAseq | -1.9196 | 0.0000 | |
| SRP193095 | PLXNA2 | 5362 | RNAseq | -1.0030 | 0.0000 | |
| SRP219564 | PLXNA2 | 5362 | RNAseq | -0.9162 | 0.0292 | |
| TCGA | PLXNA2 | 5362 | RNAseq | -0.1967 | 0.0223 |
Upregulated datasets: 0; Downregulated datasets: 6.
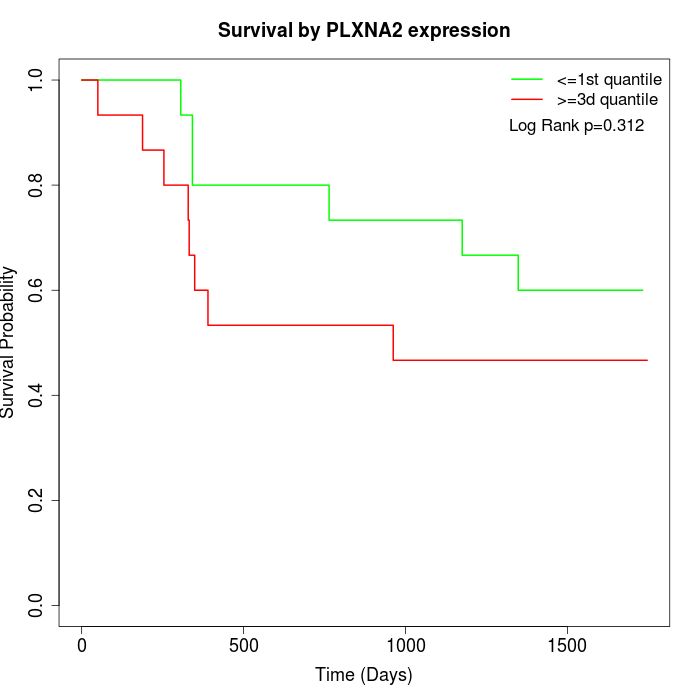
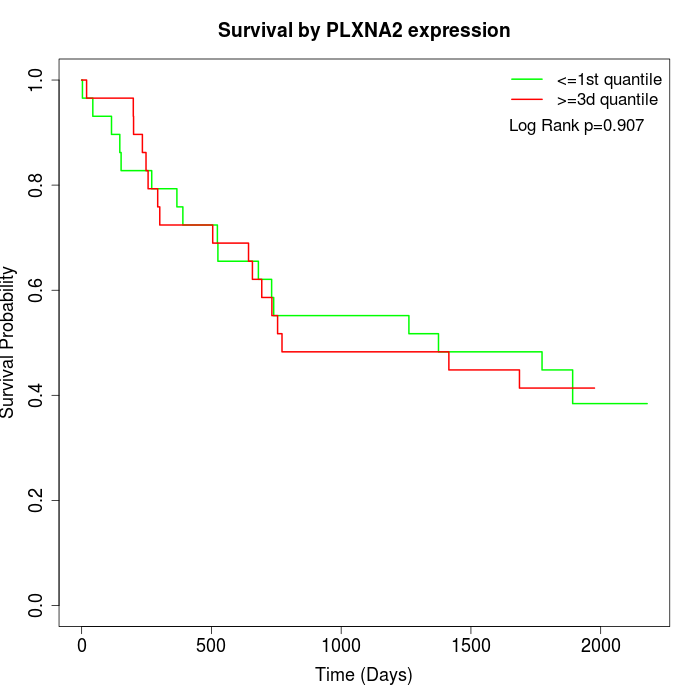
Survival by PLXNA2 expression:
Note: Click image to view full size file.
Copy number change of PLXNA2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PLXNA2 | 5362 | 11 | 0 | 19 | |
| GSE20123 | PLXNA2 | 5362 | 11 | 0 | 19 | |
| GSE43470 | PLXNA2 | 5362 | 8 | 0 | 35 | |
| GSE46452 | PLXNA2 | 5362 | 3 | 1 | 55 | |
| GSE47630 | PLXNA2 | 5362 | 14 | 0 | 26 | |
| GSE54993 | PLXNA2 | 5362 | 0 | 6 | 64 | |
| GSE54994 | PLXNA2 | 5362 | 14 | 0 | 39 | |
| GSE60625 | PLXNA2 | 5362 | 0 | 0 | 11 | |
| GSE74703 | PLXNA2 | 5362 | 8 | 0 | 28 | |
| GSE74704 | PLXNA2 | 5362 | 5 | 0 | 15 | |
| TCGA | PLXNA2 | 5362 | 45 | 4 | 47 |
Total number of gains: 119; Total number of losses: 11; Total Number of normals: 358.
Somatic mutations of PLXNA2:
Generating mutation plots.
Highly correlated genes for PLXNA2:
Showing top 20/1078 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PLXNA2 | CA13 | 0.796196 | 3 | 0 | 3 |
| PLXNA2 | PTPN23 | 0.791849 | 3 | 0 | 3 |
| PLXNA2 | ALKBH8 | 0.767044 | 3 | 0 | 3 |
| PLXNA2 | SLC10A5 | 0.764717 | 3 | 0 | 3 |
| PLXNA2 | RBM20 | 0.756307 | 3 | 0 | 3 |
| PLXNA2 | TRERF1 | 0.750684 | 3 | 0 | 3 |
| PLXNA2 | ANKRD13A | 0.740482 | 4 | 0 | 4 |
| PLXNA2 | STXBP5 | 0.737991 | 5 | 0 | 5 |
| PLXNA2 | SRP68 | 0.737835 | 3 | 0 | 3 |
| PLXNA2 | SLC46A2 | 0.736404 | 5 | 0 | 5 |
| PLXNA2 | ZNF124 | 0.735492 | 3 | 0 | 3 |
| PLXNA2 | CCNDBP1 | 0.727149 | 4 | 0 | 3 |
| PLXNA2 | CTDSP1 | 0.72642 | 3 | 0 | 3 |
| PLXNA2 | TCEANC2 | 0.723542 | 3 | 0 | 3 |
| PLXNA2 | SARS2 | 0.722324 | 3 | 0 | 3 |
| PLXNA2 | MTA3 | 0.721955 | 4 | 0 | 3 |
| PLXNA2 | CASC3 | 0.720442 | 3 | 0 | 3 |
| PLXNA2 | CEACAM6 | 0.712194 | 8 | 0 | 8 |
| PLXNA2 | TMEM120A | 0.707755 | 5 | 0 | 5 |
| PLXNA2 | ZSWIM6 | 0.707533 | 4 | 0 | 4 |
For details and further investigation, click here