| Full name: complexin 1 | Alias Symbol: CPX-I | ||
| Type: protein-coding gene | Cytoband: 4p16.3 | ||
| Entrez ID: 10815 | HGNC ID: HGNC:2309 | Ensembl Gene: ENSG00000168993 | OMIM ID: 605032 |
| Drug and gene relationship at DGIdb | |||
Expression of CPLX1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CPLX1 | 10815 | 223500_at | 0.4792 | 0.6336 | |
| GSE26886 | CPLX1 | 10815 | 223500_at | 0.5773 | 0.0236 | |
| GSE45670 | CPLX1 | 10815 | 223500_at | -0.3096 | 0.3579 | |
| GSE53622 | CPLX1 | 10815 | 133744 | -0.1518 | 0.3669 | |
| GSE53624 | CPLX1 | 10815 | 133744 | -0.0099 | 0.9375 | |
| GSE63941 | CPLX1 | 10815 | 223500_at | 0.3718 | 0.3043 | |
| GSE77861 | CPLX1 | 10815 | 223500_at | 0.2130 | 0.2182 | |
| GSE97050 | CPLX1 | 10815 | A_24_P51909 | 0.1264 | 0.5742 | |
| SRP064894 | CPLX1 | 10815 | RNAseq | 0.0953 | 0.7986 | |
| SRP133303 | CPLX1 | 10815 | RNAseq | -1.0694 | 0.0023 | |
| SRP159526 | CPLX1 | 10815 | RNAseq | 0.2375 | 0.5196 | |
| SRP219564 | CPLX1 | 10815 | RNAseq | 0.9171 | 0.2218 | |
| TCGA | CPLX1 | 10815 | RNAseq | -0.6759 | 0.0007 |
Upregulated datasets: 0; Downregulated datasets: 1.
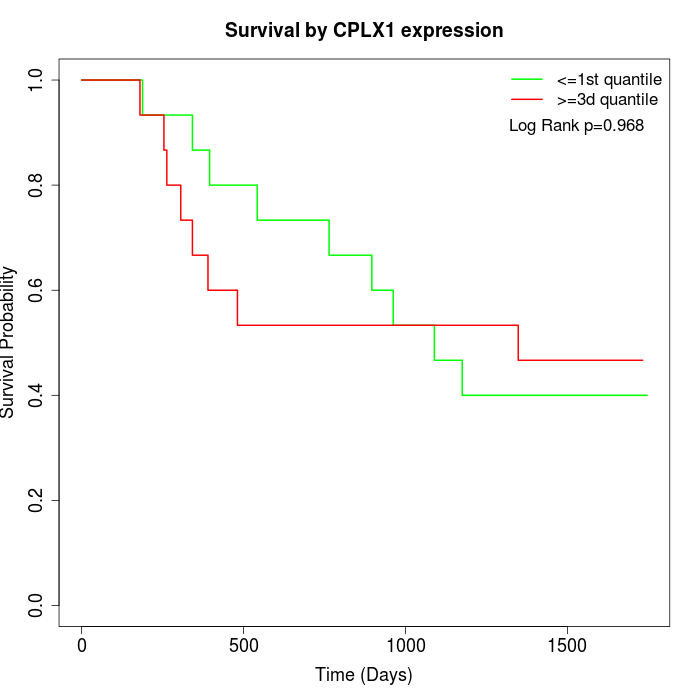
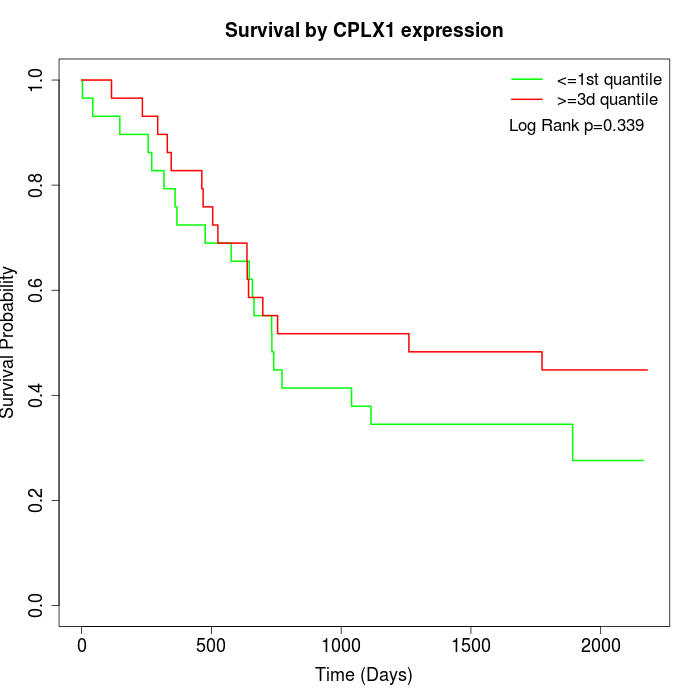
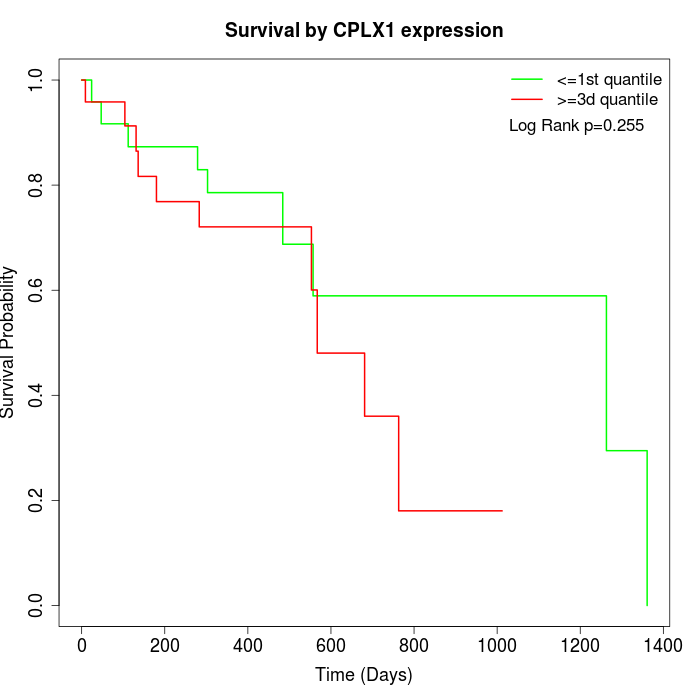
Survival by CPLX1 expression:
Note: Click image to view full size file.
Copy number change of CPLX1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CPLX1 | 10815 | 0 | 17 | 13 | |
| GSE20123 | CPLX1 | 10815 | 0 | 15 | 15 | |
| GSE43470 | CPLX1 | 10815 | 0 | 17 | 26 | |
| GSE46452 | CPLX1 | 10815 | 1 | 36 | 22 | |
| GSE47630 | CPLX1 | 10815 | 3 | 19 | 18 | |
| GSE54993 | CPLX1 | 10815 | 11 | 0 | 59 | |
| GSE54994 | CPLX1 | 10815 | 3 | 12 | 38 | |
| GSE60625 | CPLX1 | 10815 | 0 | 0 | 11 | |
| GSE74703 | CPLX1 | 10815 | 0 | 13 | 23 | |
| GSE74704 | CPLX1 | 10815 | 0 | 8 | 12 | |
| TCGA | CPLX1 | 10815 | 9 | 47 | 40 |
Total number of gains: 27; Total number of losses: 184; Total Number of normals: 277.
Somatic mutations of CPLX1:
Generating mutation plots.
Highly correlated genes for CPLX1:
Showing top 20/234 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CPLX1 | ZMIZ2 | 0.824483 | 3 | 0 | 3 |
| CPLX1 | DUSP23 | 0.802665 | 3 | 0 | 3 |
| CPLX1 | TEX9 | 0.792084 | 3 | 0 | 3 |
| CPLX1 | PTOV1 | 0.778738 | 3 | 0 | 3 |
| CPLX1 | ZNF7 | 0.774839 | 3 | 0 | 3 |
| CPLX1 | ZNF417 | 0.77228 | 3 | 0 | 3 |
| CPLX1 | POU2F1 | 0.767713 | 3 | 0 | 3 |
| CPLX1 | DNHD1 | 0.760389 | 4 | 0 | 3 |
| CPLX1 | TPP2 | 0.759407 | 3 | 0 | 3 |
| CPLX1 | GSK3A | 0.758483 | 3 | 0 | 3 |
| CPLX1 | POLR3D | 0.758101 | 3 | 0 | 3 |
| CPLX1 | ZNF518B | 0.757422 | 3 | 0 | 3 |
| CPLX1 | GZF1 | 0.756905 | 3 | 0 | 3 |
| CPLX1 | CCDC97 | 0.756883 | 3 | 0 | 3 |
| CPLX1 | MAP2K7 | 0.756443 | 3 | 0 | 3 |
| CPLX1 | LENG8 | 0.751887 | 3 | 0 | 3 |
| CPLX1 | C17orf67 | 0.75078 | 3 | 0 | 3 |
| CPLX1 | ABCD4 | 0.74386 | 3 | 0 | 3 |
| CPLX1 | ZNF713 | 0.74363 | 3 | 0 | 3 |
| CPLX1 | PIN4 | 0.741454 | 3 | 0 | 3 |
For details and further investigation, click here