| Full name: mitogen-activated protein kinase kinase 7 | Alias Symbol: MKK7|Jnkk2 | ||
| Type: protein-coding gene | Cytoband: 19p13.2 | ||
| Entrez ID: 5609 | HGNC ID: HGNC:6847 | Ensembl Gene: ENSG00000076984 | OMIM ID: 603014 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
MAP2K7 involved pathways:
Expression of MAP2K7:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MAP2K7 | 5609 | 226053_at | 0.3907 | 0.4009 | |
| GSE20347 | MAP2K7 | 5609 | 216206_x_at | 0.1212 | 0.5248 | |
| GSE23400 | MAP2K7 | 5609 | 209951_s_at | -0.2475 | 0.0000 | |
| GSE26886 | MAP2K7 | 5609 | 226023_at | 0.3267 | 0.0268 | |
| GSE29001 | MAP2K7 | 5609 | 216206_x_at | 0.2331 | 0.2987 | |
| GSE38129 | MAP2K7 | 5609 | 216206_x_at | 0.0171 | 0.9397 | |
| GSE45670 | MAP2K7 | 5609 | 226023_at | 0.0519 | 0.6019 | |
| GSE53622 | MAP2K7 | 5609 | 25084 | -0.0190 | 0.7741 | |
| GSE53624 | MAP2K7 | 5609 | 25084 | 0.3850 | 0.0000 | |
| GSE63941 | MAP2K7 | 5609 | 226053_at | 1.1311 | 0.0013 | |
| GSE77861 | MAP2K7 | 5609 | 226023_at | -0.0368 | 0.8642 | |
| GSE97050 | MAP2K7 | 5609 | A_33_P3236651 | -0.1263 | 0.5947 | |
| SRP007169 | MAP2K7 | 5609 | RNAseq | -0.3233 | 0.3910 | |
| SRP008496 | MAP2K7 | 5609 | RNAseq | -0.3362 | 0.1726 | |
| SRP064894 | MAP2K7 | 5609 | RNAseq | 0.3640 | 0.0426 | |
| SRP133303 | MAP2K7 | 5609 | RNAseq | -0.0784 | 0.5836 | |
| SRP159526 | MAP2K7 | 5609 | RNAseq | 0.0876 | 0.6274 | |
| SRP193095 | MAP2K7 | 5609 | RNAseq | 0.1752 | 0.1568 | |
| SRP219564 | MAP2K7 | 5609 | RNAseq | 0.7556 | 0.1104 | |
| TCGA | MAP2K7 | 5609 | RNAseq | -0.0015 | 0.9749 |
Upregulated datasets: 1; Downregulated datasets: 0.
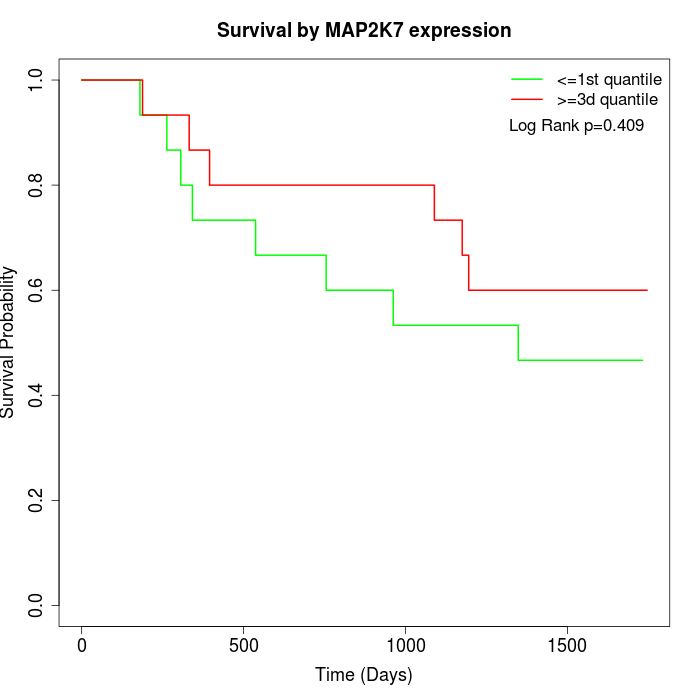
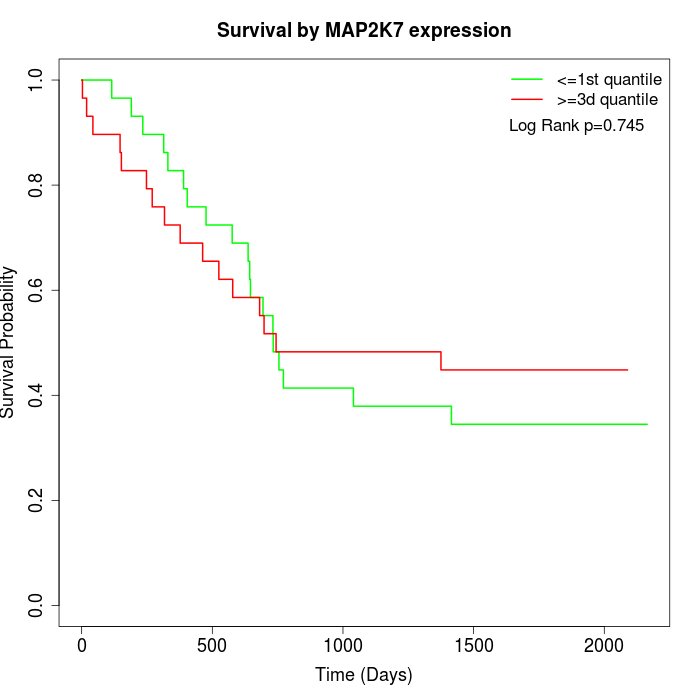
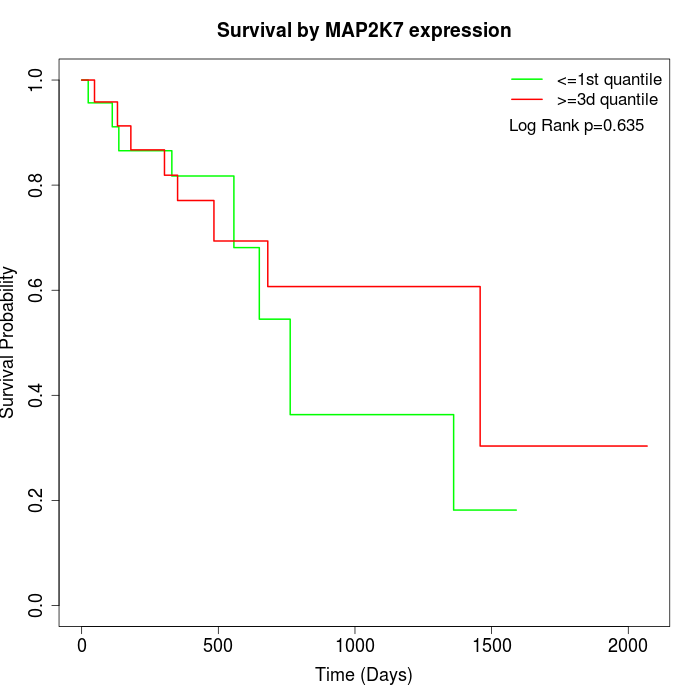
Survival by MAP2K7 expression:
Note: Click image to view full size file.
Copy number change of MAP2K7:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MAP2K7 | 5609 | 4 | 3 | 23 | |
| GSE20123 | MAP2K7 | 5609 | 3 | 2 | 25 | |
| GSE43470 | MAP2K7 | 5609 | 2 | 7 | 34 | |
| GSE46452 | MAP2K7 | 5609 | 47 | 1 | 11 | |
| GSE47630 | MAP2K7 | 5609 | 5 | 7 | 28 | |
| GSE54993 | MAP2K7 | 5609 | 16 | 3 | 51 | |
| GSE54994 | MAP2K7 | 5609 | 6 | 14 | 33 | |
| GSE60625 | MAP2K7 | 5609 | 9 | 0 | 2 | |
| GSE74703 | MAP2K7 | 5609 | 2 | 4 | 30 | |
| GSE74704 | MAP2K7 | 5609 | 0 | 2 | 18 | |
| TCGA | MAP2K7 | 5609 | 9 | 20 | 67 |
Total number of gains: 103; Total number of losses: 63; Total Number of normals: 322.
Somatic mutations of MAP2K7:
Generating mutation plots.
Highly correlated genes for MAP2K7:
Showing top 20/512 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MAP2K7 | HAUS2 | 0.872958 | 5 | 0 | 5 |
| MAP2K7 | GATAD2B | 0.813484 | 3 | 0 | 3 |
| MAP2K7 | MZT2B | 0.793332 | 6 | 0 | 5 |
| MAP2K7 | MYNN | 0.781769 | 3 | 0 | 3 |
| MAP2K7 | QSOX2 | 0.774486 | 3 | 0 | 3 |
| MAP2K7 | CRTC2 | 0.769289 | 4 | 0 | 3 |
| MAP2K7 | SURF6 | 0.757917 | 4 | 0 | 4 |
| MAP2K7 | CPLX1 | 0.756443 | 3 | 0 | 3 |
| MAP2K7 | TAS2R45 | 0.756344 | 3 | 0 | 3 |
| MAP2K7 | ZNF292 | 0.754016 | 3 | 0 | 3 |
| MAP2K7 | ERN2 | 0.7468 | 5 | 0 | 5 |
| MAP2K7 | SLC25A19 | 0.746022 | 3 | 0 | 3 |
| MAP2K7 | PREB | 0.741976 | 3 | 0 | 3 |
| MAP2K7 | NUFIP1 | 0.739358 | 3 | 0 | 3 |
| MAP2K7 | B3GAT2 | 0.73878 | 3 | 0 | 3 |
| MAP2K7 | KLHL28 | 0.736634 | 4 | 0 | 3 |
| MAP2K7 | TMEM201 | 0.728862 | 3 | 0 | 3 |
| MAP2K7 | DHDH | 0.727397 | 3 | 0 | 3 |
| MAP2K7 | FOXC1 | 0.727116 | 3 | 0 | 3 |
| MAP2K7 | ZXDC | 0.727093 | 4 | 0 | 4 |
For details and further investigation, click here