| Full name: cysteine rich protein 3 | Alias Symbol: TLP-A|bA480N24.2|TLP | ||
| Type: protein-coding gene | Cytoband: 6p21.1 | ||
| Entrez ID: 401262 | HGNC ID: HGNC:17751 | Ensembl Gene: ENSG00000146215 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of CRIP3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CRIP3 | 401262 | 235720_at | 0.1220 | 0.6040 | |
| GSE26886 | CRIP3 | 401262 | 235720_at | 0.2397 | 0.0259 | |
| GSE45670 | CRIP3 | 401262 | 235720_at | -0.0128 | 0.9253 | |
| GSE53622 | CRIP3 | 401262 | 61590 | -0.3658 | 0.0267 | |
| GSE53624 | CRIP3 | 401262 | 7273 | -0.5674 | 0.0000 | |
| GSE63941 | CRIP3 | 401262 | 235720_at | 0.0607 | 0.7368 | |
| GSE77861 | CRIP3 | 401262 | 235720_at | -0.1363 | 0.2038 | |
| TCGA | CRIP3 | 401262 | RNAseq | 0.6441 | 0.4242 |
Upregulated datasets: 0; Downregulated datasets: 0.
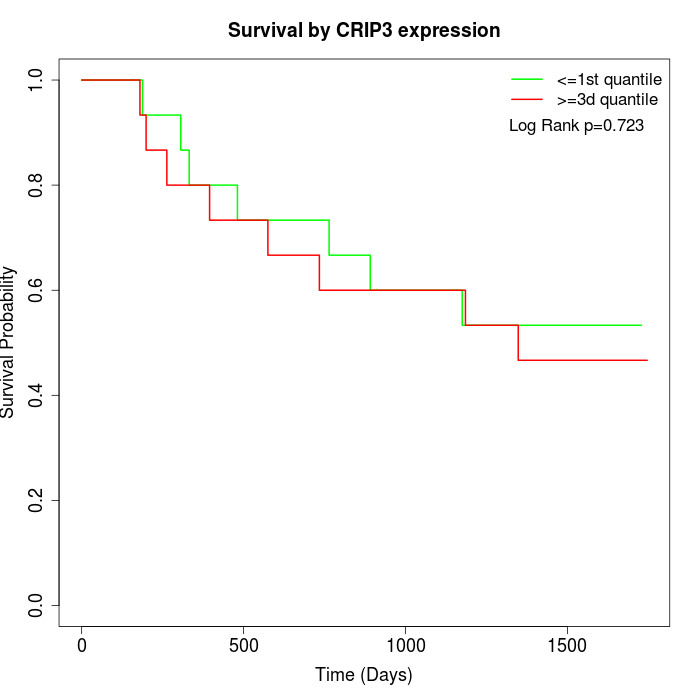
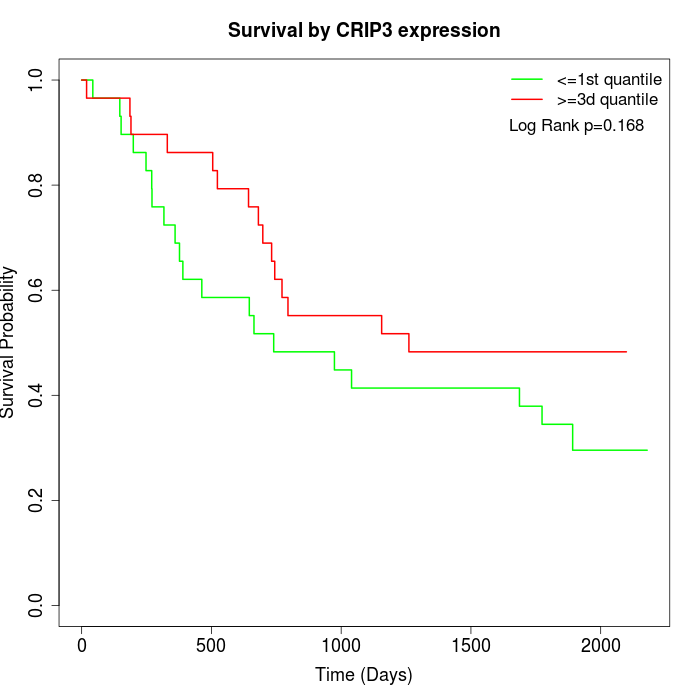
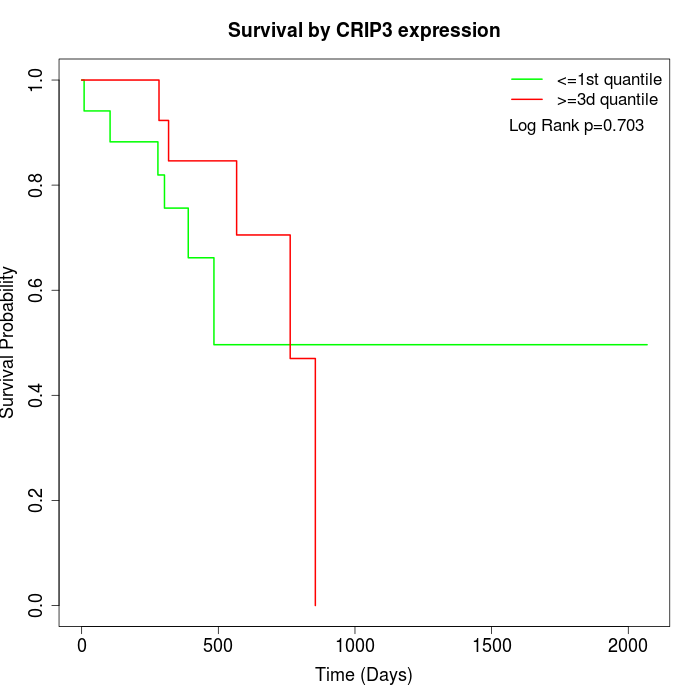
Survival by CRIP3 expression:
Note: Click image to view full size file.
Copy number change of CRIP3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CRIP3 | 401262 | 7 | 0 | 23 | |
| GSE20123 | CRIP3 | 401262 | 7 | 0 | 23 | |
| GSE43470 | CRIP3 | 401262 | 6 | 0 | 37 | |
| GSE46452 | CRIP3 | 401262 | 2 | 9 | 48 | |
| GSE47630 | CRIP3 | 401262 | 8 | 4 | 28 | |
| GSE54993 | CRIP3 | 401262 | 3 | 2 | 65 | |
| GSE54994 | CRIP3 | 401262 | 11 | 4 | 38 | |
| GSE60625 | CRIP3 | 401262 | 0 | 1 | 10 | |
| GSE74703 | CRIP3 | 401262 | 6 | 0 | 30 | |
| GSE74704 | CRIP3 | 401262 | 3 | 0 | 17 | |
| TCGA | CRIP3 | 401262 | 20 | 13 | 63 |
Total number of gains: 73; Total number of losses: 33; Total Number of normals: 382.
Somatic mutations of CRIP3:
Generating mutation plots.
Highly correlated genes for CRIP3:
Showing top 20/59 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CRIP3 | TSC22D4 | 0.742536 | 3 | 0 | 3 |
| CRIP3 | ZSWIM4 | 0.734068 | 3 | 0 | 3 |
| CRIP3 | FABP2 | 0.727671 | 3 | 0 | 3 |
| CRIP3 | ZNF653 | 0.724866 | 3 | 0 | 3 |
| CRIP3 | DLG4 | 0.698194 | 3 | 0 | 3 |
| CRIP3 | NFAM1 | 0.697486 | 3 | 0 | 3 |
| CRIP3 | TRIM51 | 0.687286 | 3 | 0 | 3 |
| CRIP3 | KHDC3L | 0.686411 | 3 | 0 | 3 |
| CRIP3 | ELL | 0.676627 | 3 | 0 | 3 |
| CRIP3 | VCX2 | 0.668759 | 3 | 0 | 3 |
| CRIP3 | TEX11 | 0.664876 | 3 | 0 | 3 |
| CRIP3 | HMX1 | 0.657383 | 3 | 0 | 3 |
| CRIP3 | PEX11G | 0.654173 | 3 | 0 | 3 |
| CRIP3 | TMEM132E | 0.65235 | 3 | 0 | 3 |
| CRIP3 | KCNC3 | 0.651658 | 3 | 0 | 3 |
| CRIP3 | CPTP | 0.64028 | 3 | 0 | 3 |
| CRIP3 | ASPDH | 0.628599 | 4 | 0 | 3 |
| CRIP3 | AIRE | 0.621663 | 4 | 0 | 3 |
| CRIP3 | GPR162 | 0.620851 | 3 | 0 | 3 |
| CRIP3 | FKBP6 | 0.61808 | 3 | 0 | 3 |
For details and further investigation, click here