| Full name: crystallin gamma B | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 2q33.3 | ||
| Entrez ID: 1419 | HGNC ID: HGNC:2409 | Ensembl Gene: ENSG00000182187 | OMIM ID: 123670 |
| Drug and gene relationship at DGIdb | |||
Expression of CRYGB:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CRYGB | 1419 | 207715_at | 0.1366 | 0.5342 | |
| GSE20347 | CRYGB | 1419 | 207715_at | -0.0408 | 0.5155 | |
| GSE23400 | CRYGB | 1419 | 207715_at | -0.0826 | 0.0092 | |
| GSE26886 | CRYGB | 1419 | 207715_at | 0.1751 | 0.2804 | |
| GSE29001 | CRYGB | 1419 | 207715_at | -0.2844 | 0.0571 | |
| GSE38129 | CRYGB | 1419 | 207715_at | -0.0768 | 0.2857 | |
| GSE45670 | CRYGB | 1419 | 207715_at | 0.0788 | 0.3990 | |
| GSE53622 | CRYGB | 1419 | 69307 | 0.3649 | 0.1232 | |
| GSE53624 | CRYGB | 1419 | 69307 | 0.2482 | 0.0091 | |
| GSE63941 | CRYGB | 1419 | 207715_at | 0.0209 | 0.8878 | |
| GSE77861 | CRYGB | 1419 | 207715_at | 0.0096 | 0.9399 |
Upregulated datasets: 0; Downregulated datasets: 0.
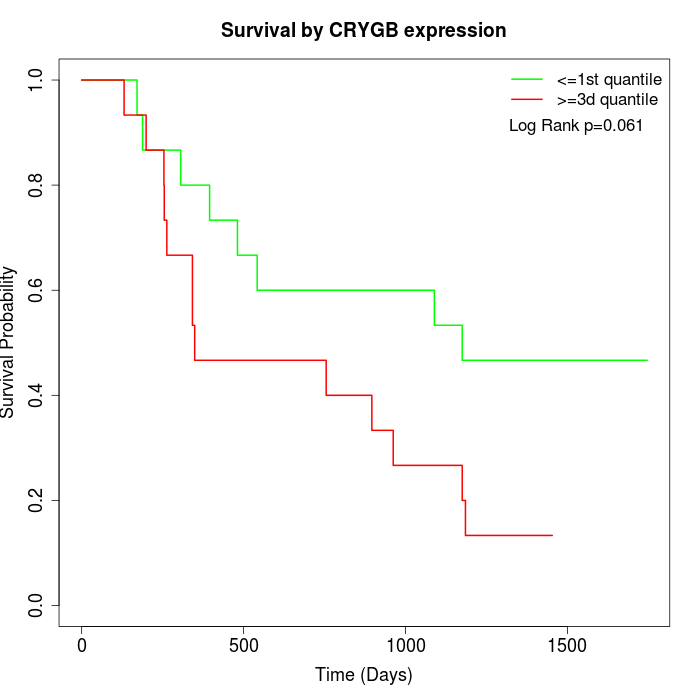
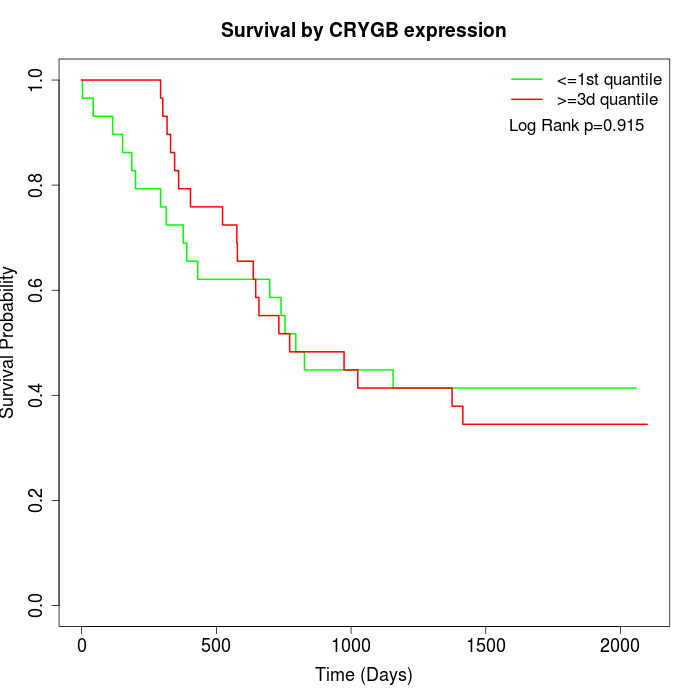
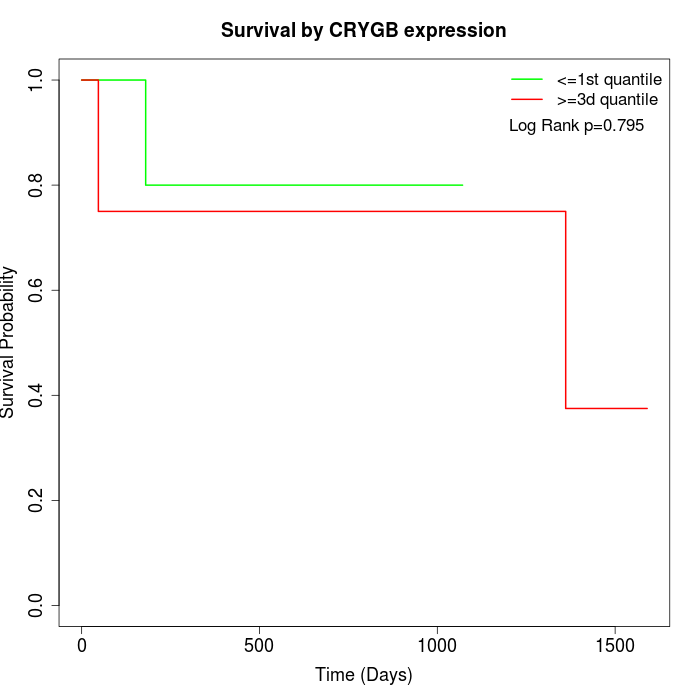
Survival by CRYGB expression:
Note: Click image to view full size file.
Copy number change of CRYGB:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CRYGB | 1419 | 3 | 10 | 17 | |
| GSE20123 | CRYGB | 1419 | 3 | 10 | 17 | |
| GSE43470 | CRYGB | 1419 | 2 | 4 | 37 | |
| GSE46452 | CRYGB | 1419 | 0 | 5 | 54 | |
| GSE47630 | CRYGB | 1419 | 4 | 5 | 31 | |
| GSE54993 | CRYGB | 1419 | 0 | 3 | 67 | |
| GSE54994 | CRYGB | 1419 | 9 | 9 | 35 | |
| GSE60625 | CRYGB | 1419 | 0 | 3 | 8 | |
| GSE74703 | CRYGB | 1419 | 2 | 3 | 31 | |
| GSE74704 | CRYGB | 1419 | 2 | 5 | 13 | |
| TCGA | CRYGB | 1419 | 15 | 20 | 61 |
Total number of gains: 40; Total number of losses: 77; Total Number of normals: 371.
Somatic mutations of CRYGB:
Generating mutation plots.
Highly correlated genes for CRYGB:
Showing top 20/578 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CRYGB | ZNF835 | 0.724768 | 3 | 0 | 3 |
| CRYGB | TEX11 | 0.69988 | 5 | 0 | 5 |
| CRYGB | HTR6 | 0.682838 | 5 | 0 | 5 |
| CRYGB | CACNA1H | 0.675926 | 3 | 0 | 3 |
| CRYGB | PSG9 | 0.675912 | 6 | 0 | 6 |
| CRYGB | SPTBN4 | 0.674312 | 4 | 0 | 4 |
| CRYGB | MYOD1 | 0.664377 | 3 | 0 | 3 |
| CRYGB | TLL2 | 0.663893 | 4 | 0 | 4 |
| CRYGB | AP2A1 | 0.660973 | 3 | 0 | 3 |
| CRYGB | CAMKV | 0.658217 | 3 | 0 | 3 |
| CRYGB | ARHGEF38 | 0.654223 | 4 | 0 | 3 |
| CRYGB | KRT76 | 0.650337 | 3 | 0 | 3 |
| CRYGB | NEUROD6 | 0.646185 | 4 | 0 | 3 |
| CRYGB | CCR10 | 0.6458 | 3 | 0 | 3 |
| CRYGB | SERINC2 | 0.645486 | 5 | 0 | 5 |
| CRYGB | RHEBL1 | 0.644258 | 3 | 0 | 3 |
| CRYGB | ART4 | 0.644237 | 5 | 0 | 5 |
| CRYGB | BTNL2 | 0.642609 | 6 | 0 | 6 |
| CRYGB | CPLX3 | 0.641935 | 4 | 0 | 4 |
| CRYGB | OR11A1 | 0.63977 | 4 | 0 | 3 |
For details and further investigation, click here